Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants
Marine Lefebvre, Henri Chahinian, Bernard La Scola, Jacques Fantini
Viruses, doi:10.3390/v16121836
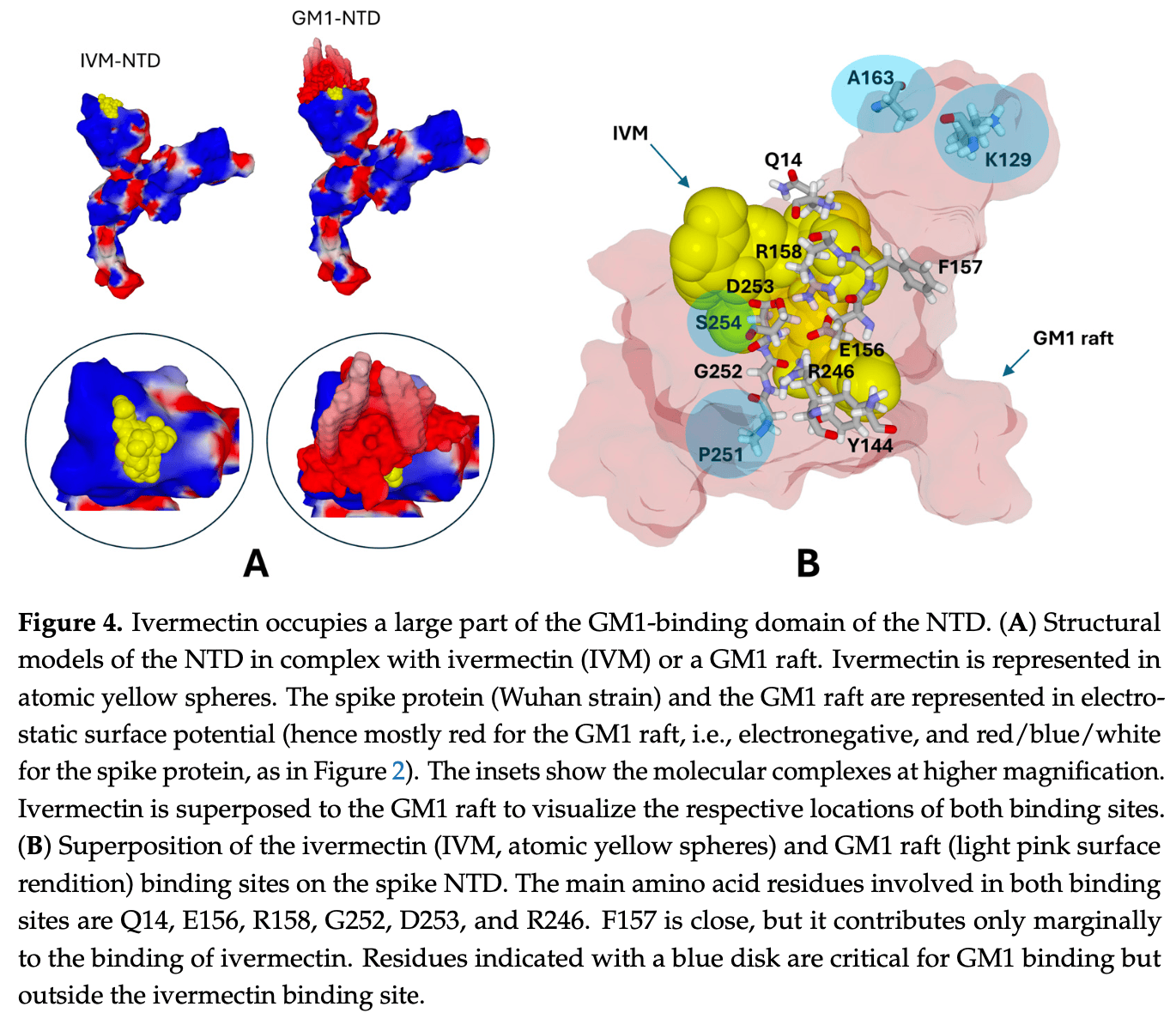
Most studies on the docking of ivermectin on the spike protein of SARS-CoV-2 concern the receptor binding domain (RBD) and, more precisely, the RBD interface recognized by the ACE2 receptor. The N-terminal domain (NTD), which controls the initial attachment of the virus to lipid raft gangliosides, has not received the attention it deserves. In this study, we combined molecular modeling and physicochemical approaches to analyze the mode of interaction of ivermectin with the interface of the NTD-facing lipid rafts of the host cell membrane. We characterize a binding area that presents point mutations and deletions in successive SARS-CoV-2 variants from the initial strain to omicron KP.3 circulating in many countries in 2024. We show that ivermectin has exceptional flexibility, allowing the drug to bind to the spike protein of all variants tested. The energy of interaction is specific to each variant, allowing a classification according to their affinity for ivermectin in the following ascending order: Omicron KP.3 < Delta < Omicron BA.5 < Alpha < Wuhan (B.1) < Omicron BA.1. The binding site of ivermectin is subject to important variations of the NTD, including the Y144 deletion. It overlaps with the ganglioside binding domain of the NTD, as demonstrated by docking and physicochemical studies. These results suggest a new mechanism of antiviral action for ivermectin based on competitive inhibition for initial virus attachment to lipid rafts. The current KP.3 variant is still recognized by ivermectin, although with an affinity slightly lower than the Wuhan strain.
Conflicts of Interest: The authors declare no conflicts of interest. Viruses 2024, 16, 1836
References
Allen, Introduction to molecular dynamics simulation, Comput. Soft Matter Synth. Polym. Proteins
Alonso, Bliznyuk, Gready, Combining docking and molecular dynamic simulations in drug design, Med. Res. Rev,
doi:10.1002/med.20067Amanat, Thapa, Lei, Ahmed, Adelsberg et al., SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2, Cell,
doi:10.1016/j.cell.2021.06.005Aminpour, Cannariato, Preto, Safaeeardebili, Moracchiato et al., In silico analysis of the multi-targeted mode of action of ivermectin and related compounds, Computation,
doi:10.3390/computation10040051Andreani, Le Bideau, Duflot, Jardot, Rolland et al., In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect, Microb. Pathog,
doi:10.1016/j.micpath.2020.104228Andrei, Conjugate Gradient Methods
Ayodele, Bamigbade, Bamigbade, Adeniyi, Tachin et al., Illustrated Procedure to Perform Molecular Docking Using PyRx and Biovia Discovery Studio Visualizer: A Case Study of 10kt With Atropine, Prog. Drug Discov. Biomed. Sci,
doi:10.36877/pddbs.a0000424Borcik, Eason, Yekefallah, Amani, Han et al., A cholesterol dimer stabilizes the inactivated state of an inward-rectifier Potassium Channel, Angew. Chem. Int. Ed,
doi:10.1002/anie.202112232Borges-Araújo, Patmanidis, Singh, Santos, Sieradzan et al., Pragmatic coarse-graining of proteins: Models and applications, J. Chem. Theory Comput,
doi:10.1021/acs.jctc.3c00733Boschi, Scheim, Bancod, Militello, Bideau et al., SARS-CoV-2 spike protein induces hemagglutination: Implications for COVID-19 morbidities and therapeutics and for vaccine adverse effects, Int. J. Mol. Sci,
doi:10.3390/ijms232415480Buttenschoen, Morris, Deane, Posebusters, AI-based docking methods fail to generate physically valid poses or generalise to novel sequences, Chem. Sci,
doi:10.1039/D3SC04185AChangeux, Edelstein, Conformational selection or induced fit? 50 years of debate resolved, Biol. Rep,
doi:10.3410/B3-19Choudhury, Das, Patra, Bhattacharya, Ghosh et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: An in silico approach, Future Virol,
doi:10.2217/fvl-2020-0342Di Scala, Fantini, Hybrid In Silico/In Vitro Approaches for the Identification of Functional Cholesterol-Binding Domains in Membrane Proteins, Methods Mol. Biol,
doi:10.1007/978-1-4939-6875-6_2Di Scala, Troadec, Lelièvre, Garmy, Fantini et al., Mechanism of cholesterol-assisted oligomeric channel formation by a short Alzheimer β-amyloid peptide, J. Neurochem,
doi:10.1111/jnc.12390Dima, Salvagno, Lippi, Effects of recombinant SARS-CoV-2 spike protein variants on red blood cells parameters and red blood cell distribution width, Biomed. J,
doi:10.1016/j.bj.2024.100787Diociaiuti, Giordani, Kamel, Brasili, Sennato et al., Monosialoganglioside-GM1 triggers binding of the amyloid-protein salmon calcitonin to a Langmuir membrane model mimicking the occurrence of lipid-rafts, Biochem. Biophys. Rep,
doi:10.1016/j.bbrep.2016.10.005Ewing, Makino, Skillman, Kuntz, Dock, 4.0: Search strategies for automated molecular docking of flexible molecule databases, J. Comput.-Aided Mol. Des,
doi:10.1023/A:1011115820450Fantini, Azzaz, Chahinian, Yahi, Electrostatic Surface Potential as a Key Parameter in Virus Transmission and Evolution: How to Manage Future Virus Pandemics in the Post-COVID-19 Era, Viruses,
doi:10.3390/v15020284Fantini, Chahinian, Yahi, Convergent Evolution Dynamics of SARS-CoV-2 and HIV Surface Envelope Glycoproteins Driven by Host Cell Surface Receptors and Lipid Rafts: Lessons for the Future, Int. J. Mol. Sci,
doi:10.3390/ijms24031923Fantini, Chahinian, Yahi, Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19, Biochem. Biophys. Res. Commun,
doi:10.1016/j.bbrc.2020.10.015Fantini, Chahinian, Yahi, Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: What molecular dynamics studies of virus-host interactions reveal, Int. J. Antimicrob. Agents,
doi:10.1016/j.ijantimicag.2020.106020Fantini, Di Scala, Chahinian, Yahi, Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection, Int. J. Antimicrob. Agents,
doi:10.1016/j.ijantimicag.2020.105960Fantini, Fundamental Mechanisms in Membrane Receptology: Old Paradigms, New Concepts and Perspectives, Receptors,
doi:10.3390/receptors3010006Fantini, Yahi, Azzaz, Chahinian, Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating Covid-19 outbreaks, J. Infect,
doi:10.1016/j.jinf.2021.06.001Fantini, Yahi, Colson, Chahinian, La Scola et al., The puzzling mutational landscape of the SARS-2-variant Omicron, J. Med. Virol,
doi:10.1002/jmv.27577Gaetano, Capasso, Delre, Pirone, Saviano et al., More Is Always Better Than One: The N-Terminal Domain of the Spike Protein as Another Emerging Target for Hampering the SARS-CoV-2 Attachment to Host Cells, Int. J. Mol. Sci,
doi:10.3390/ijms22126462Ghoula, Naceri, Sitruk, Flatters, Moroy et al., Identifying promising druggable binding sites and their flexibility to target the receptor-binding domain of SARS-CoV-2 spike protein, Comput. Struct. Biotechnol. J,
doi:10.1016/j.csbj.2023.03.029Grippo, Lucidi, A globally convergent version of the Polak-Ribiere conjugate gradient method, Math. Program,
doi:10.1007/BF02614362Guex, Peitsch, SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling, Electrophoresis,
doi:10.1002/elps.1150181505Gyselinck, Janssens, Verhamme, Vos, Rationale for azithromycin in COVID-19: An overview of existing evidence, BMJ Open Respir. Res,
doi:10.1136/bmjresp-2020-000806Huang, Kalyanaraman, Bernacki, Jacobson, Molecular mechanics methods for predicting protein-ligand binding, Phys. Chem. Chem. Phys. PCCP,
doi:10.1039/B608269FHuang, Wong, Wheeler, Flexible protein-flexible ligand docking with disrupted velocity simulated annealing, Proteins,
doi:10.1002/prot.21781Islas, Scior, Allosteric Binding of MDMA to the Human Serotonin Transporter (hSERT) via Ensemble Binding Space Analysis with ∆G Calculations, Induced Fit Docking and Monte Carlo Simulations, Molecules,
doi:10.3390/molecules27092977Jaafar, Boschi, Aherfi, Bancod, Le Bideau et al., High individual heterogeneity of neutralizing activities against the original strain and nine different variants of SARS-CoV-2, Viruses,
doi:10.3390/v13112177Kaku, Yo, Tolentino, Uriu, Okumura et al., Virological characteristics of the SARS-CoV-2 KP. 3, LB. 1, and KP. 2.3 variants, Lancet Infect. Dis,
doi:10.1016/S1473-3099(24)00415-8Kharche, Sengupta, Dynamic protein interfaces and conformational landscapes of membrane protein complexes, Curr. Opin. Struct. Biol,
doi:10.1016/j.sbi.2020.01.001Kim, Cha, Choi, Kim, Omega class glutathione S-transferase: Antioxidant enzyme in pathogenesis of neurodegenerative diseases, Oxidative Med. Cell. Longev,
doi:10.1155/2017/5049532Kim, Chivian, Baker, Protein structure prediction and analysis using the Robetta server, Nucleic Acids Res,
doi:10.1093/nar/gkh468Klatzmann, Mcdougal, Maddon, The CD4 molecule and HIV infection, Immunodefic. Rev
Lan, Ge, Yu, Shan, Zhou et al., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor, Nature,
doi:10.1038/s41586-020-2180-5Liu, Bakker, Narimatsu, Van Kuppeveld, Clausen et al., H3N2 influenza A virus gradually adapts to human-type receptor binding and entry specificity after the start of the 1968 pandemic, Proc. Natl. Acad. Sci,
doi:10.1073/pnas.2304992120Low, Yip, Lal, Repositioning Ivermectin for Covid-19 treatment: Molecular mechanisms of action against SARS-CoV-2 replication, Biochim. Biophys. Acta-Mol. Basis Dis,
doi:10.1016/j.bbadis.2021.166294Maresca, Derghal, Caravagna, Dudin, Fantini, Controlled aggregation of adenine by sugars: Physicochemical studies, molecular modelling simulations of sugar-aromatic CH-pi stacking interactions, and biological significance, Phys. Chem. Chem. Phys. PCCP,
doi:10.1039/b802594kMatveeva, Lefebvre, Chahinian, Yahi, Fantini, Host membranes as drivers of virus evolution, Viruses,
doi:10.3390/v15091854Mcdougal, Maddon, Dalgleish, Clapham, Littman et al., The T4 glycoprotein is a cell-surface receptor for the AIDS virus, Cold Spring Harb. Symp. Quant. Biol,
doi:10.1101/SQB.1986.051.01.083Milanetti, Miotto, Di Rienzo, Nagaraj, Monti et al., In-silico evidence for a two receptor based strategy of SARS-CoV-2, Front. Mol. Biosci,
doi:10.3389/fmolb.2021.690655Monti, Milanetti, Frans, Miotto, Di Rienzo et al., Two Receptor Binding Strategy of SARS-CoV-2 Is Mediated by Both the N-Terminal and Receptor-Binding Spike Domain, J. Phys. Chem. B,
doi:10.1021/acs.jpcb.3c06258Nocedal, Wright, Conjugate gradient methods
Sarkar, Concilio, Sessa, Marrafino, Piotto, Advancements and novel approaches in modified autodock vina algorithms for enhanced molecular docking, Results Chem,
doi:10.1016/j.rechem.2024.101319Scheim, A deadly embrace: Hemagglutination mediated by SARS-CoV-2 spike protein at its 22 N-glycosylation sites, red blood cell surface sialoglycoproteins, and antibody, Int. J. Mol. Sci,
doi:10.3390/ijms23052558Scheim, Vottero, Santin, Hirsh, Sialylated glycan bindings from SARS-CoV-2 spike protein to blood and endothelial cells govern the severe morbidities of COVID-19, Int. J. Mol. Sci,
doi:10.3390/ijms242317039Seyran, Takayama, Uversky, Lundstrom, Palù et al., The structural basis of accelerated host cell entry by SARS-CoV-2, FEBS J,
doi:10.1111/febs.15651Sokkar, Mohandass, Ramachandran, Multiple templates-based homology modeling enhances structure quality of AT1 receptor: Validation by molecular dynamics and antagonist docking, J. Mol. Model,
doi:10.1007/s00894-010-0860-zSrinivasu, Babu, Rao, Energy Minimization of CDK2 bound ligands: A Computational Approach, Int. J. Eng. Res. Appl
Ströh, Nagarathinam, Krey, Conformational flexibility in the CD81-binding site of the hepatitis C virus glycoprotein E2, Front. Immunol,
doi:10.3389/fimmu.2018.01396Suryadevara, Shrihari, Gilchuk, Vanblargan, Binshtein et al., Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein, Cell,
doi:10.1016/j.cell.2021.03.029Thomsen, Christensen, Moldock, A new technique for high-accuracy molecular docking, J. Med. Chem,
doi:10.1021/jm051197eTrott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem,
doi:10.1002/jcc.21334Tsuji, Docking Study with HyperChem, Revision G1
Tsuji, Homology Modeling Professional for HyperChem
Tsuji, Shudo, Kagechika, Docking simulations suggest that all-trans retinoic acid could bind to retinoid X receptors, J. Comput.-Aided Mol. Des,
doi:10.1007/s10822-015-9869-9Uversky, Intrinsically disordered proteins and their environment: Effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding, Protein J,
doi:10.1007/s10930-009-9201-4Wlodarski, Zagrovic, Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin, Proc. Natl. Acad. Sci,
doi:10.1073/pnas.0906966106DOI record:
{
"DOI": "10.3390/v16121836",
"ISSN": [
"1999-4915"
],
"URL": "http://dx.doi.org/10.3390/v16121836",
"abstract": "<jats:p>Most studies on the docking of ivermectin on the spike protein of SARS-CoV-2 concern the receptor binding domain (RBD) and, more precisely, the RBD interface recognized by the ACE2 receptor. The N-terminal domain (NTD), which controls the initial attachment of the virus to lipid raft gangliosides, has not received the attention it deserves. In this study, we combined molecular modeling and physicochemical approaches to analyze the mode of interaction of ivermectin with the interface of the NTD-facing lipid rafts of the host cell membrane. We characterize a binding area that presents point mutations and deletions in successive SARS-CoV-2 variants from the initial strain to omicron KP.3 circulating in many countries in 2024. We show that ivermectin has exceptional flexibility, allowing the drug to bind to the spike protein of all variants tested. The energy of interaction is specific to each variant, allowing a classification according to their affinity for ivermectin in the following ascending order: Omicron KP.3 < Delta < Omicron BA.5 < Alpha < Wuhan (B.1) < Omicron BA.1. The binding site of ivermectin is subject to important variations of the NTD, including the Y144 deletion. It overlaps with the ganglioside binding domain of the NTD, as demonstrated by docking and physicochemical studies. These results suggest a new mechanism of antiviral action for ivermectin based on competitive inhibition for initial virus attachment to lipid rafts. The current KP.3 variant is still recognized by ivermectin, although with an affinity slightly lower than the Wuhan strain.</jats:p>",
"alternative-id": [
"v16121836"
],
"author": [
{
"affiliation": [
{
"name": "IHU Méditerranée Infection, 19-21 Boulevard Jean Moulin, 13005 Marseille, France"
},
{
"name": "Microbes Evolution Phylogeny and Infections (MEPHI), Aix-Marseille Université, 27 Boulevard Jean Moulin, 13005 Marseille, France"
},
{
"name": "Assistance Publique-Hôpitaux de Marseille (AP-HM), 264 Rue Saint-Pierre, 13005 Marseille, France"
},
{
"name": "Department of Biology, Faculty of Medicine, Aix-Marseille University, INSERM UA16, 13015 Marseille, France"
}
],
"family": "Lefebvre",
"given": "Marine",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-9516-4168",
"affiliation": [
{
"name": "Department of Biology, Faculty of Medicine, Aix-Marseille University, INSERM UA16, 13015 Marseille, France"
}
],
"authenticated-orcid": false,
"family": "Chahinian",
"given": "Henri",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8006-7704",
"affiliation": [
{
"name": "IHU Méditerranée Infection, 19-21 Boulevard Jean Moulin, 13005 Marseille, France"
},
{
"name": "Microbes Evolution Phylogeny and Infections (MEPHI), Aix-Marseille Université, 27 Boulevard Jean Moulin, 13005 Marseille, France"
},
{
"name": "Assistance Publique-Hôpitaux de Marseille (AP-HM), 264 Rue Saint-Pierre, 13005 Marseille, France"
}
],
"authenticated-orcid": false,
"family": "La Scola",
"given": "Bernard",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8653-5521",
"affiliation": [
{
"name": "Department of Biology, Faculty of Medicine, Aix-Marseille University, INSERM UA16, 13015 Marseille, France"
}
],
"authenticated-orcid": false,
"family": "Fantini",
"given": "Jacques",
"sequence": "additional"
}
],
"container-title": "Viruses",
"container-title-short": "Viruses",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
11,
27
]
],
"date-time": "2024-11-27T09:20:12Z",
"timestamp": 1732699212000
},
"deposited": {
"date-parts": [
[
2024,
11,
27
]
],
"date-time": "2024-11-27T09:37:24Z",
"timestamp": 1732700244000
},
"indexed": {
"date-parts": [
[
2024,
11,
28
]
],
"date-time": "2024-11-28T05:23:07Z",
"timestamp": 1732771387549,
"version": "3.29.0"
},
"is-referenced-by-count": 0,
"issue": "12",
"issued": {
"date-parts": [
[
2024,
11,
27
]
]
},
"journal-issue": {
"issue": "12",
"published-online": {
"date-parts": [
[
2024,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
27
]
],
"date-time": "2024-11-27T00:00:00Z",
"timestamp": 1732665600000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1999-4915/16/12/1836/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1836",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2024,
11,
27
]
]
},
"published-online": {
"date-parts": [
[
2024,
11,
27
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1101/SQB.1986.051.01.083",
"article-title": "The T4 glycoprotein is a cell-surface receptor for the AIDS virus",
"author": "McDougal",
"doi-asserted-by": "crossref",
"first-page": "703",
"journal-title": "Cold Spring Harb. Symp. Quant. Biol.",
"key": "ref_1",
"volume": "51",
"year": "1986"
},
{
"article-title": "The CD4 molecule and HIV infection",
"author": "Klatzmann",
"first-page": "43",
"journal-title": "Immunodefic. Rev.",
"key": "ref_2",
"volume": "2",
"year": "1990"
},
{
"DOI": "10.1038/s41586-020-2179-y",
"article-title": "Structural basis of receptor recognition by SARS-CoV-2",
"author": "Shang",
"doi-asserted-by": "crossref",
"first-page": "221",
"journal-title": "Nature",
"key": "ref_3",
"volume": "581",
"year": "2020"
},
{
"DOI": "10.1016/j.jmb.2018.06.024",
"article-title": "Virus–Receptor Interactions: The Key to Cellular Invasion",
"author": "Maginnis",
"doi-asserted-by": "crossref",
"first-page": "2590",
"journal-title": "J. Mol. Biol.",
"key": "ref_4",
"volume": "430",
"year": "2018"
},
{
"DOI": "10.3390/v15020284",
"doi-asserted-by": "crossref",
"key": "ref_5",
"unstructured": "Fantini, J., Azzaz, F., Chahinian, H., and Yahi, N. (2023). Electrostatic Surface Potential as a Key Parameter in Virus Transmission and Evolution: How to Manage Future Virus Pandemics in the Post-COVID-19 Era. Viruses, 15."
},
{
"DOI": "10.3390/v15091854",
"doi-asserted-by": "crossref",
"key": "ref_6",
"unstructured": "Matveeva, M., Lefebvre, M., Chahinian, H., Yahi, N., and Fantini, J. (2023). Host membranes as drivers of virus evolution. Viruses, 15."
},
{
"DOI": "10.3390/ijms24031923",
"doi-asserted-by": "crossref",
"key": "ref_7",
"unstructured": "Fantini, J., Chahinian, H., and Yahi, N. (2023). Convergent Evolution Dynamics of SARS-CoV-2 and HIV Surface Envelope Glycoproteins Driven by Host Cell Surface Receptors and Lipid Rafts: Lessons for the Future. Int. J. Mol. Sci., 24."
},
{
"DOI": "10.3389/fmolb.2021.690655",
"doi-asserted-by": "crossref",
"key": "ref_8",
"unstructured": "Milanetti, E., Miotto, M., Di Rienzo, L., Nagaraj, M., Monti, M., Golbek, T.W., Gosti, G., Roeters, S.J., Weidner, T., and Otzen, D.E. (2021). In-silico evidence for a two receptor based strategy of SARS-CoV-2. Front. Mol. Biosci., 8."
},
{
"DOI": "10.1021/acs.jpcb.3c06258",
"article-title": "Two Receptor Binding Strategy of SARS-CoV-2 Is Mediated by Both the N-Terminal and Receptor-Binding Spike Domain",
"author": "Monti",
"doi-asserted-by": "crossref",
"first-page": "451",
"journal-title": "J. Phys. Chem. B",
"key": "ref_9",
"volume": "128",
"year": "2024"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105960",
"article-title": "Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "105960",
"journal-title": "Int. J. Antimicrob. Agents",
"key": "ref_10",
"volume": "55",
"year": "2020"
},
{
"DOI": "10.1016/j.bbrc.2020.10.015",
"article-title": "Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "132",
"journal-title": "Biochem. Biophys. Res. Commun.",
"key": "ref_11",
"volume": "538",
"year": "2021"
},
{
"DOI": "10.1111/febs.15651",
"article-title": "The structural basis of accelerated host cell entry by SARS-CoV-2",
"author": "Seyran",
"doi-asserted-by": "crossref",
"first-page": "5010",
"journal-title": "FEBS J.",
"key": "ref_12",
"volume": "288",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2180-5",
"article-title": "Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor",
"author": "Lan",
"doi-asserted-by": "crossref",
"first-page": "215",
"journal-title": "Nature",
"key": "ref_13",
"volume": "581",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2021.03.029",
"article-title": "Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein",
"author": "Suryadevara",
"doi-asserted-by": "crossref",
"first-page": "2316",
"journal-title": "Cell",
"key": "ref_14",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2021.06.005",
"article-title": "SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2",
"author": "Amanat",
"doi-asserted-by": "crossref",
"first-page": "3936",
"journal-title": "Cell",
"key": "ref_15",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.3390/v13112177",
"doi-asserted-by": "crossref",
"key": "ref_16",
"unstructured": "Jaafar, R., Boschi, C., Aherfi, S., Bancod, A., Le Bideau, M., Edouard, S., Colson, P., Chahinian, H., Raoult, D., and Yahi, N. (2021). High individual heterogeneity of neutralizing activities against the original strain and nine different variants of SARS-CoV-2. Viruses, 13."
},
{
"DOI": "10.1016/j.ijantimicag.2020.106020",
"article-title": "Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: What molecular dynamics studies of virus-host interactions reveal",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "106020",
"journal-title": "Int. J. Antimicrob. Agents",
"key": "ref_17",
"volume": "56",
"year": "2020"
},
{
"DOI": "10.1136/bmjresp-2020-000806",
"article-title": "Rationale for azithromycin in COVID-19: An overview of existing evidence",
"author": "Gyselinck",
"doi-asserted-by": "crossref",
"first-page": "e000806",
"journal-title": "BMJ Open Respir. Res.",
"key": "ref_18",
"volume": "8",
"year": "2021"
},
{
"DOI": "10.1016/j.micpath.2020.104228",
"article-title": "In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect",
"author": "Andreani",
"doi-asserted-by": "crossref",
"first-page": "104228",
"journal-title": "Microb. Pathog.",
"key": "ref_19",
"volume": "145",
"year": "2020"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"article-title": "The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro",
"author": "Caly",
"doi-asserted-by": "crossref",
"first-page": "104787",
"journal-title": "Antivir. Res.",
"key": "ref_20",
"volume": "178",
"year": "2020"
},
{
"DOI": "10.1016/j.bbadis.2021.166294",
"doi-asserted-by": "crossref",
"key": "ref_21",
"unstructured": "Low, Z.Y., Yip, A.J.W., and Lal, S.K. (2022). Repositioning Ivermectin for Covid-19 treatment: Molecular mechanisms of action against SARS-CoV-2 replication. Biochim. Biophys. Acta-Mol. Basis Dis., 1868."
},
{
"DOI": "10.1080/23744235.2024.2385500",
"article-title": "Features of the SARS-CoV-2 KP. 3 variant mutations",
"author": "Branda",
"doi-asserted-by": "crossref",
"first-page": "894",
"journal-title": "Infect. Dis.",
"key": "ref_22",
"volume": "56",
"year": "2024"
},
{
"DOI": "10.1016/S1473-3099(24)00298-6",
"article-title": "Virological characteristics of the SARS-CoV-2 KP. 2 variant",
"author": "Kaku",
"doi-asserted-by": "crossref",
"first-page": "e416",
"journal-title": "Lancet Infect. Dis.",
"key": "ref_23",
"volume": "24",
"year": "2024"
},
{
"DOI": "10.1016/S1473-3099(24)00415-8",
"article-title": "Virological characteristics of the SARS-CoV-2 KP. 3, LB. 1, and KP. 2.3 variants",
"author": "Kaku",
"doi-asserted-by": "crossref",
"first-page": "e482",
"journal-title": "Lancet Infect. Dis.",
"key": "ref_24",
"volume": "24",
"year": "2024"
},
{
"DOI": "10.4103/apjtm.apjtm_341_24",
"article-title": "Ever-evolving SARS-CoV-2: Latest variant KP. 2 is on the rise",
"author": "Shanmugaraj",
"doi-asserted-by": "crossref",
"first-page": "241",
"journal-title": "Asian Pac. J. Trop. Med.",
"key": "ref_25",
"volume": "17",
"year": "2024"
},
{
"DOI": "10.1016/j.jinf.2021.06.001",
"article-title": "Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating Covid-19 outbreaks",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "197",
"journal-title": "J. Infect.",
"key": "ref_26",
"volume": "83",
"year": "2021"
},
{
"DOI": "10.1093/nar/gkh468",
"article-title": "Protein structure prediction and analysis using the Robetta server",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "W526",
"journal-title": "Nucleic Acids Res.",
"key": "ref_27",
"volume": "32",
"year": "2004"
},
{
"DOI": "10.1002/elps.1150181505",
"article-title": "SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling",
"author": "Guex",
"doi-asserted-by": "crossref",
"first-page": "2714",
"journal-title": "Electrophoresis",
"key": "ref_28",
"volume": "18",
"year": "1997"
},
{
"DOI": "10.1155/2017/5049532",
"article-title": "Omega class glutathione S-transferase: Antioxidant enzyme in pathogenesis of neurodegenerative diseases",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "5049532",
"journal-title": "Oxidative Med. Cell. Longev.",
"key": "ref_29",
"volume": "2017",
"year": "2017"
},
{
"key": "ref_30",
"unstructured": "Tsuji, M. (2015). Docking Study with HyperChem, Revision G1, Institute of Molecular Function."
},
{
"DOI": "10.1007/s10822-015-9869-9",
"article-title": "Docking simulations suggest that all-trans retinoic acid could bind to retinoid X receptors",
"author": "Tsuji",
"doi-asserted-by": "crossref",
"first-page": "975",
"journal-title": "J. Comput.-Aided Mol. Des.",
"key": "ref_31",
"volume": "29",
"year": "2015"
},
{
"key": "ref_32",
"unstructured": "Tsuji, M. (2015). Homology Modeling Professional for HyperChem, Revision G1, Institute of Molecular Function."
},
{
"article-title": "Energy Minimization of CDK2 bound ligands: A Computational Approach",
"author": "Srinivasu",
"first-page": "1884",
"journal-title": "Int. J. Eng. Res. Appl.",
"key": "ref_33",
"volume": "2",
"year": "2012"
},
{
"DOI": "10.1021/jm051197e",
"article-title": "MolDock: A new technique for high-accuracy molecular docking",
"author": "Thomsen",
"doi-asserted-by": "crossref",
"first-page": "3315",
"journal-title": "J. Med. Chem.",
"key": "ref_34",
"volume": "49",
"year": "2006"
},
{
"DOI": "10.1016/j.colsurfb.2009.07.043",
"article-title": "Surface chemistry of Alzheimer’s disease: A Langmuir monolayer approach",
"author": "Thakur",
"doi-asserted-by": "crossref",
"first-page": "436",
"journal-title": "Colloids Surf. B Biointerfaces",
"key": "ref_35",
"volume": "74",
"year": "2009"
},
{
"DOI": "10.1016/j.chemphyslip.2018.01.008",
"article-title": "Langmuir-monolayer methodologies for characterizing protein-lipid interactions",
"author": "Elderdfi",
"doi-asserted-by": "crossref",
"first-page": "61",
"journal-title": "Chem. Phys. Lipids",
"key": "ref_36",
"volume": "212",
"year": "2018"
},
{
"article-title": "Monosialoganglioside-GM1 triggers binding of the amyloid-protein salmon calcitonin to a Langmuir membrane model mimicking the occurrence of lipid-rafts",
"author": "Diociaiuti",
"first-page": "365",
"journal-title": "Biochem. Biophys. Rep.",
"key": "ref_37",
"volume": "8",
"year": "2016"
},
{
"DOI": "10.1007/978-1-4939-6875-6_2",
"article-title": "Hybrid In Silico/In Vitro Approaches for the Identification of Functional Cholesterol-Binding Domains in Membrane Proteins",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "7",
"journal-title": "Methods Mol. Biol.",
"key": "ref_38",
"volume": "1583",
"year": "2017"
},
{
"DOI": "10.36877/pddbs.a0000424",
"article-title": "Illustrated Procedure to Perform Molecular Docking Using PyRx and Biovia Discovery Studio Visualizer: A Case Study of 10kt With Atropine",
"author": "Ayodele",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Prog. Drug Discov. Biomed. Sci.",
"key": "ref_39",
"volume": "6",
"year": "2023"
},
{
"DOI": "10.1023/A:1011115820450",
"article-title": "DOCK 4.0: Search strategies for automated molecular docking of flexible molecule databases",
"author": "Ewing",
"doi-asserted-by": "crossref",
"first-page": "411",
"journal-title": "J. Comput.-Aided Mol. Des.",
"key": "ref_40",
"volume": "15",
"year": "2001"
},
{
"DOI": "10.1002/jcc.21334",
"article-title": "AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading",
"author": "Trott",
"doi-asserted-by": "crossref",
"first-page": "455",
"journal-title": "J. Comput. Chem.",
"key": "ref_41",
"volume": "31",
"year": "2010"
},
{
"DOI": "10.1146/annurev-biochem-030222-120000",
"article-title": "The Art and Science of Molecular Docking",
"author": "Paggi",
"doi-asserted-by": "crossref",
"first-page": "389",
"journal-title": "Annu. Rev. Biochem.",
"key": "ref_42",
"volume": "93",
"year": "2024"
},
{
"DOI": "10.1016/j.tips.2014.12.001",
"article-title": "Beware of docking!",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "78",
"journal-title": "Trends Pharmacol. Sci.",
"key": "ref_43",
"volume": "36",
"year": "2015"
},
{
"DOI": "10.1039/D3SC04185A",
"article-title": "PoseBusters: AI-based docking methods fail to generate physically valid poses or generalise to novel sequences",
"author": "Buttenschoen",
"doi-asserted-by": "crossref",
"first-page": "3130",
"journal-title": "Chem. Sci.",
"key": "ref_44",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1016/j.rechem.2024.101319",
"article-title": "Advancements and novel approaches in modified autodock vina algorithms for enhanced molecular docking",
"author": "Sarkar",
"doi-asserted-by": "crossref",
"first-page": "101319",
"journal-title": "Results Chem.",
"key": "ref_45",
"volume": "7",
"year": "2024"
},
{
"DOI": "10.1002/prot.21781",
"article-title": "Flexible protein-flexible ligand docking with disrupted velocity simulated annealing",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "440",
"journal-title": "Proteins",
"key": "ref_46",
"volume": "71",
"year": "2008"
},
{
"DOI": "10.3410/B3-19",
"doi-asserted-by": "crossref",
"key": "ref_47",
"unstructured": "Changeux, J.-P., and Edelstein, S. (2011). Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep., 3."
},
{
"DOI": "10.1016/j.bpc.2014.03.003",
"article-title": "Distinguishing induced fit from conformational selection",
"author": "Gianni",
"doi-asserted-by": "crossref",
"first-page": "33",
"journal-title": "Biophys. Chem.",
"key": "ref_48",
"volume": "189",
"year": "2014"
},
{
"DOI": "10.1073/pnas.0906966106",
"article-title": "Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin",
"author": "Wlodarski",
"doi-asserted-by": "crossref",
"first-page": "19346",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_49",
"volume": "106",
"year": "2009"
},
{
"DOI": "10.3390/molecules27092977",
"doi-asserted-by": "crossref",
"key": "ref_50",
"unstructured": "Islas, Á.A., and Scior, T. (2022). Allosteric Binding of MDMA to the Human Serotonin Transporter (hSERT) via Ensemble Binding Space Analysis with ΔG Calculations, Induced Fit Docking and Monte Carlo Simulations. Molecules, 27."
},
{
"DOI": "10.1007/s10930-009-9201-4",
"article-title": "Intrinsically disordered proteins and their environment: Effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding",
"author": "Uversky",
"doi-asserted-by": "crossref",
"first-page": "305",
"journal-title": "Protein J.",
"key": "ref_51",
"volume": "28",
"year": "2009"
},
{
"DOI": "10.1517/17460441.2012.686489",
"article-title": "Intrinsically disordered proteins and novel strategies for drug discovery",
"author": "Uversky",
"doi-asserted-by": "crossref",
"first-page": "475",
"journal-title": "Expert Opin. Drug Discov.",
"key": "ref_52",
"volume": "7",
"year": "2012"
},
{
"DOI": "10.1021/cr500288y",
"article-title": "Introduction to intrinsically disordered proteins (IDPs)",
"author": "Uversky",
"doi-asserted-by": "crossref",
"first-page": "6557",
"journal-title": "Chem. Rev.",
"key": "ref_53",
"volume": "114",
"year": "2014"
},
{
"DOI": "10.1039/B608269F",
"article-title": "Molecular mechanics methods for predicting protein-ligand binding",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "5166",
"journal-title": "Phys. Chem. Chem. Phys. PCCP",
"key": "ref_54",
"volume": "8",
"year": "2006"
},
{
"article-title": "Introduction to molecular dynamics simulation",
"author": "Allen",
"first-page": "1",
"journal-title": "Comput. Soft Matter Synth. Polym. Proteins",
"key": "ref_55",
"volume": "23",
"year": "2004"
},
{
"DOI": "10.1007/s00894-010-0860-z",
"article-title": "Multiple templates-based homology modeling enhances structure quality of AT1 receptor: Validation by molecular dynamics and antagonist docking",
"author": "Sokkar",
"doi-asserted-by": "crossref",
"first-page": "1565",
"journal-title": "J. Mol. Model.",
"key": "ref_56",
"volume": "17",
"year": "2011"
},
{
"DOI": "10.1016/j.ejmech.2014.08.004",
"article-title": "Molecular dynamics in drug design",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "4",
"journal-title": "Eur. J. Med. Chem.",
"key": "ref_57",
"volume": "91",
"year": "2015"
},
{
"DOI": "10.1002/med.20067",
"article-title": "Combining docking and molecular dynamic simulations in drug design",
"author": "Alonso",
"doi-asserted-by": "crossref",
"first-page": "531",
"journal-title": "Med. Res. Rev.",
"key": "ref_58",
"volume": "26",
"year": "2006"
},
{
"DOI": "10.1002/anie.202112232",
"article-title": "A cholesterol dimer stabilizes the inactivated state of an inward-rectifier Potassium Channel",
"author": "Borcik",
"doi-asserted-by": "crossref",
"first-page": "e202112232",
"journal-title": "Angew. Chem. Int. Ed.",
"key": "ref_59",
"volume": "61",
"year": "2022"
},
{
"DOI": "10.3390/membranes12090844",
"doi-asserted-by": "crossref",
"key": "ref_60",
"unstructured": "Sinha, S., Tam, B., and Wang, S.M. (2022). Applications of molecular dynamics simulation in protein study. Membranes, 12."
},
{
"DOI": "10.1016/j.sbi.2020.01.001",
"article-title": "Dynamic protein interfaces and conformational landscapes of membrane protein complexes",
"author": "Kharche",
"doi-asserted-by": "crossref",
"first-page": "191",
"journal-title": "Curr. Opin. Struct. Biol.",
"key": "ref_61",
"volume": "61",
"year": "2020"
},
{
"DOI": "10.1021/acs.jctc.3c00733",
"article-title": "Pragmatic coarse-graining of proteins: Models and applications",
"author": "Patmanidis",
"doi-asserted-by": "crossref",
"first-page": "7112",
"journal-title": "J. Chem. Theory Comput.",
"key": "ref_62",
"volume": "19",
"year": "2023"
},
{
"DOI": "10.1007/978-0-387-40065-5_5",
"doi-asserted-by": "crossref",
"key": "ref_63",
"unstructured": "Nocedal, J., and Wright, S.J. (2006). Conjugate gradient methods. Numerical Optimization, Springer."
},
{
"DOI": "10.1007/978-3-031-08720-2_5",
"doi-asserted-by": "crossref",
"key": "ref_64",
"unstructured": "Andrei, N. (2022). Conjugate Gradient Methods. Modern Numerical Nonlinear Optimization, Springer International Publishing."
},
{
"DOI": "10.1007/BF02614362",
"article-title": "A globally convergent version of the Polak-Ribiere conjugate gradient method",
"author": "Grippo",
"doi-asserted-by": "crossref",
"first-page": "375",
"journal-title": "Math. Program.",
"key": "ref_65",
"volume": "78",
"year": "1997"
},
{
"DOI": "10.1007/BF00941472",
"article-title": "Generalized Polak-Ribiere algorithm",
"author": "Khoda",
"doi-asserted-by": "crossref",
"first-page": "345",
"journal-title": "J. Optim. Theory Appl.",
"key": "ref_66",
"volume": "75",
"year": "1992"
},
{
"DOI": "10.1039/b802594k",
"article-title": "Controlled aggregation of adenine by sugars: Physicochemical studies, molecular modelling simulations of sugar-aromatic CH-pi stacking interactions, and biological significance",
"author": "Maresca",
"doi-asserted-by": "crossref",
"first-page": "2792",
"journal-title": "Phys. Chem. Chem. Phys. PCCP",
"key": "ref_67",
"volume": "10",
"year": "2008"
},
{
"DOI": "10.1107/S090744490402356X",
"article-title": "Introduction to macromolecular refinement",
"author": "Tronrud",
"doi-asserted-by": "crossref",
"first-page": "2156",
"journal-title": "Acta Crystallogr. Sect. D Biol. Crystallogr.",
"key": "ref_68",
"volume": "60",
"year": "2004"
},
{
"DOI": "10.1111/jnc.12390",
"article-title": "Mechanism of cholesterol-assisted oligomeric channel formation by a short Alzheimer β-amyloid peptide",
"author": "Troadec",
"doi-asserted-by": "crossref",
"first-page": "186",
"journal-title": "J. Neurochem.",
"key": "ref_69",
"volume": "128",
"year": "2014"
},
{
"DOI": "10.2217/fvl-2020-0342",
"article-title": "Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: An in silico approach",
"author": "Choudhury",
"doi-asserted-by": "crossref",
"first-page": "277",
"journal-title": "Future Virol.",
"key": "ref_70",
"volume": "16",
"year": "2021"
},
{
"DOI": "10.3390/computation10040051",
"doi-asserted-by": "crossref",
"key": "ref_71",
"unstructured": "Aminpour, M., Cannariato, M., Preto, J., Safaeeardebili, M.E., Moracchiato, A., Doria, D., Donato, F., Zizzi, E.A., Deriu, M.A., and Scheim, D.E. (2022). In silico analysis of the multi-targeted mode of action of ivermectin and related compounds. Computation, 10."
},
{
"DOI": "10.1093/glycob/cwab032",
"article-title": "The role of cell surface sialic acids for SARS-CoV-2 infection",
"author": "Sun",
"doi-asserted-by": "crossref",
"first-page": "1245",
"journal-title": "Glycobiology",
"key": "ref_72",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.3390/ijms22126462",
"doi-asserted-by": "crossref",
"key": "ref_73",
"unstructured": "Di Gaetano, S., Capasso, D., Delre, P., Pirone, L., Saviano, M., Pedone, E., and Mangiatordi, G.F. (2021). More Is Always Better Than One: The N-Terminal Domain of the Spike Protein as Another Emerging Target for Hampering the SARS-CoV-2 Attachment to Host Cells. Int. J. Mol. Sci., 22."
},
{
"DOI": "10.1101/2022.11.24.517882",
"doi-asserted-by": "crossref",
"key": "ref_74",
"unstructured": "Boschi, C., Scheim, D.E., Bancod, A., Militello, M., Bideau, M.L., Colson, P., Fantini, J., and Scola, B.L. (2022). SARS-CoV-2 spike protein induces hemagglutination: Implications for COVID-19 morbidities and therapeutics and for vaccine adverse effects. Int. J. Mol. Sci., 23."
},
{
"DOI": "10.1002/jmv.27577",
"article-title": "The puzzling mutational landscape of the SARS-2-variant Omicron",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "2019",
"journal-title": "J. Med. Virol.",
"key": "ref_75",
"volume": "94",
"year": "2022"
},
{
"DOI": "10.1002/2211-5463.13612",
"article-title": "Lipid rafts and human diseases: Why we need to target gangliosides",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "1636",
"journal-title": "FEBS Open Bio",
"key": "ref_76",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.3390/receptors3010006",
"article-title": "Fundamental Mechanisms in Membrane Receptology: Old Paradigms, New Concepts and Perspectives",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "107",
"journal-title": "Receptors",
"key": "ref_77",
"volume": "3",
"year": "2024"
},
{
"DOI": "10.1073/pnas.2304992120",
"article-title": "H3N2 influenza A virus gradually adapts to human-type receptor binding and entry specificity after the start of the 1968 pandemic",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "e2304992120",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_78",
"volume": "120",
"year": "2023"
},
{
"DOI": "10.3389/fimmu.2018.01396",
"doi-asserted-by": "crossref",
"key": "ref_79",
"unstructured": "Ströh, L.J., Nagarathinam, K., and Krey, T. (2018). Conformational flexibility in the CD81-binding site of the hepatitis C virus glycoprotein E2. Front. Immunol., 9."
},
{
"DOI": "10.1016/j.csbj.2023.03.029",
"article-title": "Identifying promising druggable binding sites and their flexibility to target the receptor-binding domain of SARS-CoV-2 spike protein",
"author": "Ghoula",
"doi-asserted-by": "crossref",
"first-page": "2339",
"journal-title": "Comput. Struct. Biotechnol. J.",
"key": "ref_80",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1038/s41429-021-00491-6",
"article-title": "The mechanisms of action of ivermectin against SARS-CoV-2—An extensive review",
"author": "Zaidi",
"doi-asserted-by": "crossref",
"first-page": "60",
"journal-title": "J. Antibiot.",
"key": "ref_81",
"volume": "75",
"year": "2022"
},
{
"DOI": "10.3390/ijms23052558",
"doi-asserted-by": "crossref",
"key": "ref_82",
"unstructured": "Scheim, D.E. (2022). A deadly embrace: Hemagglutination mediated by SARS-CoV-2 spike protein at its 22 N-glycosylation sites, red blood cell surface sialoglycoproteins, and antibody. Int. J. Mol. Sci., 23."
},
{
"DOI": "10.3390/ijms242317039",
"doi-asserted-by": "crossref",
"key": "ref_83",
"unstructured": "Scheim, D.E., Vottero, P., Santin, A.D., and Hirsh, A.G. (2023). Sialylated glycan bindings from SARS-CoV-2 spike protein to blood and endothelial cells govern the severe morbidities of COVID-19. Int. J. Mol. Sci., 24."
},
{
"DOI": "10.1016/j.bj.2024.100787",
"doi-asserted-by": "crossref",
"key": "ref_84",
"unstructured": "Dima, F., Salvagno, G.L., and Lippi, G. (2024). Effects of recombinant SARS-CoV-2 spike protein variants on red blood cells parameters and red blood cell distribution width. Biomed. J., 47."
}
],
"reference-count": 84,
"references-count": 84,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1999-4915/16/12/1836"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants",
"type": "journal-article",
"volume": "16"
}