Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach
Md. Sorwer Alam Parvez, Md. Adnan Karim, Mahmudul Hasan, Jomana Jaman, Ziaul Karim, Tohura Tahsin, Md. Nazmul Hasan, Mohammad Jakir Hosen
International Journal of Biological Macromolecules, doi:10.1016/j.ijbiomac.2020.09.098
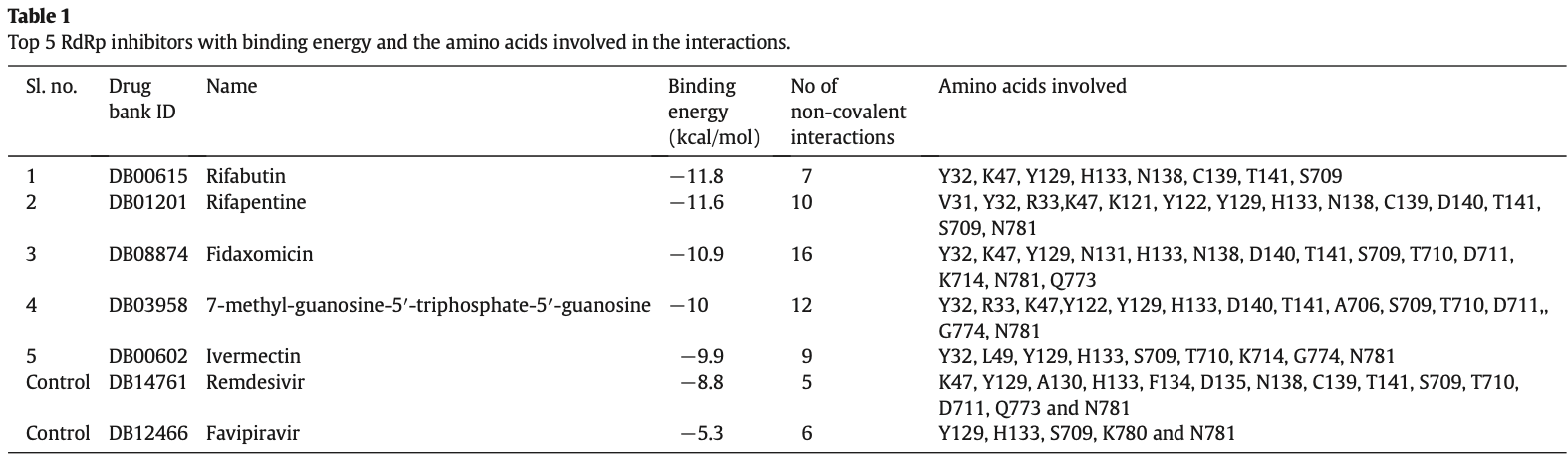
The pandemic prevalence of COVID-19 has become a very serious global health issue. Scientists all over the world have been seriously attempting in the discovery of a drug to combat SARS-CoV-2. It has been found that RNAdependent RNA polymerase (RdRp) plays a crucial role in SARS-CoV-2 replication, and thus could be a potential drug target. Here, comprehensive computational approaches including drug repurposing and molecular docking were employed to predict an effective drug candidate targeting RdRp of SARS-CoV-2. This study revealed that Rifabutin, Rifapentine, Fidaxomicin, 7-methyl-guanosine-5′-triphosphate-5′-guanosine and Ivermectin have a potential inhibitory interaction with RdRp of SARS-CoV-2 and could be effective drugs for COVID-19. In addition, virtual screening of the compounds from ZINC database also allowed the prediction of two compounds (ZINC09128258 and ZINC09883305) with pharmacophore features that interact effectively with RdRp of SARS-CoV-2, indicating their potentiality as effective inhibitors of the enzyme. Furthermore, ADME analysis along with analysis of toxicity was also undertaken to check the pharmacokinetics and drug-likeness properties of the two compounds. Comparative structural analysis of protein-inhibitor complexes revealed that the amino acids Y32, K47, Y122, Y129, H133, N138, D140, T141, S709 and N781 are crucial for drug surface hotspot in the RdRp of SARS-CoV-2.
Fig. 10 . PCA results trejectories for the protein-inhibitor complexes. Here, RdRp in complex with (A) Rifabutin, (B) ZINC09128258, and (C) ZINC098833. In this graph, the colour blue to red frames over time.
Declaration of competing interest The authors declare that they have no competing interests.
References
Agostini, Andres, Sims, Graham, Sheahan et al., Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease, MBio,
doi:10.1128/mBio.00221-18Agrawal, Singh, Srivastava, Singh, Kishore et al., Benchmarking of Different Molecular Docking Methods for Protein-Peptide Docking
Biovia, Discovery Studio Modeling Environment, Release 2017
Case, Cheatham, Darden, Gohlke, Luo et al., The Amber Biomolecular Simulation Programs,
doi:10.1002/jcc.20290Chan, Yuan, Kok, To, Chu et al., A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster, Lancet,
doi:10.1016/S0140-6736(20)30154-9Chen, Zhou, Dong, Qu, Gong et al., Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study, Lancet,
doi:10.1016/S0140-6736(20)30211-7Daina, Michielin, Zoete, SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules, Sci. Rep,
doi:10.1038/srep42717Daina, Zoete, A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules, ChemMedChem,
doi:10.1002/cmdc.201600182Delano, Pymol: an open-source molecular graphics tool, CCP4, Newsl Prot. Crystallogr
Ekins, Mestres, Testa, In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling, Br. J. Pharmacol,
doi:10.1038/sj.bjp.0707305Elfiky, Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study, Life Sci,
doi:10.1016/j.lfs.2020.117592Hsu, Chen, Lin, Yang, iGEMDOCK: A Graphical Environment of Enhancing GEMDOCK Using Pharmacological Interactions and Post-screening Analysis
Hubbard, Chen, Davis, Informatics and modeling challenges in fragmentbased drug discovery, Curr. Opin. Drug Discov. Dev
Irwin, Shoichet, ZINCa free database of commercially available compounds for virtual screening, J. Chem. Inf. Model,
doi:10.1021/ci049714+Koes, Camacho, ZINCPharmer: pharmacophore search of the ZINC database, Nucleic Acids Res,
doi:10.1093/nar/gks378Laskowski, Swindells, LigPlot+: multiple ligand-protein interaction diagrams for drug discovery, J. Chem. Inf. Model,
doi:10.1021/ci200227uLeach, Molecular Modelling: Principles and Applications
Li, Du, Wang, Wu, Li et al., In silico estimation of chemical carcinogenicity with binary and ternary classification methods, Mol. Inform,
doi:10.1002/minf.201400127Lung, Lin, Yang, Chou, Shu et al., The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase, J. Med. Virol,
doi:10.1002/jmv.25761Mishra, Dahima, In vitro ADME studies of TUG-891, a GPR-120 inhibitor using SWISS ADME predictor, J. Drug Deliv. Ther,
doi:10.22270/jddt.v9i2-s.2710Pal, Berhanu, Desalegn, Kandi, Severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2): an update, Cureus,
doi:10.7759/cureus.7423Pizzorno, Padey, Terrier, Rosa-Calatrava, Drug repurposing approaches for the treatment of influenza viral infection: reviving old drugs to fight against a longlived enemy, Front. Immunol,
doi:10.3389/fimmu.2019.00531Porcheddu, Serra, Kelvin, Kelvin, Rubino, Similarity in case fatality rates (CFR) of COVID-19/SARS-COV-2 in Italy and China, J. Infect. Dev. Count,
doi:10.3855/jidc.12600Prichard, Kern, Orthopoxvirus targets for the development of antiviral therapies, Curr. Drug Targets. Infect. Disord,
doi:10.2174/1568005053174627Rose, Prlić, Altunkaya, Bi, Bradley et al., The RCSB protein data bank: integrative view of protein, gene and 3D structural information, Nucleic Acids Res
Sakano, Takako, Iqbal Mahamood, Yamashita, Molecular dynamics analysis to evaluate docking pose prediction, Biophys. Physicobiol,
doi:10.2142/biophysico.13.0Schneidman-Duhovny, Dror, Inbar, Nussinov, Wolfson, Deterministic pharmacophore detection via multiple flexible alignment of drug-like molecules, J. Comput. Biol,
doi:10.1089/cmb.2007.0130Schneidman-Duhovny, Dror, Inbar, Nussinov, Wolfson, PharmaGist: a webserver for ligand-based pharmacophore detection, Nucleic Acids Res,
doi:10.1093/nar/gkn187Schneidman-Duhovny, Inbar, Nussinov, Wolfson, PatchDock and SymmDock : Servers for Rigid and Symmetric Docking,
doi:10.1093/nar/gki481Shen, Cheng, Xu, Li, Tang, Estimation of ADME properties with substructure pattern recognition, J. Chem. Inf. Model,
doi:10.1021/ci100104jTripathi, Ghosh, Talapatra, Bioavailability prediction of phytochemicals present in Calotropis procera (Aiton) R. Br. by using Swiss-ADME tool, World Sci. News
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem,
doi:10.1002/jcc.21334Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res,
doi:10.1038/s41422-020-0282-0Who, Coronavirus Disease (COVID-19) Situation Dashboard
Wishart, Feunang, Guo, Lo, Marcu et al., DrugBank 5.0: a major update to the DrugBank database for 2018, Nucleic Acids Res,
doi:10.1093/nar/gkx1037Xu, Cheng, Chen, Du, Li et al., In silico prediction of chemical ames mutagenicity, J. Chem. Inf. Model,
doi:10.1021/ci300400aXu, Hassounah, Colby-Germinario, Oliveira, Fogarty et al., Purification of Zika virus RNA-dependent RNA polymerase and its use to identify smallmolecule Zika inhibitors, J. Antimicrob. Chemother,
doi:10.1093/jac/dkw514Yang, Lu, Liu, Wang, Zhang et al., Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China, MedRxiv,
doi:10.1101/2020.02.10.20021675Yang, Wang, Chen, Hao, Yang, LARMD : Integration of Bioinformatic Resources to Profile Ligand-driven Protein Dynamics With a Case on the Activation of Estrogen Receptor,
doi:10.1093/bib/bbz141Zhou, Dai, Tong, COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression, J. Antimicrob. Chemother,
doi:10.1093/jac/dkaa114Zumla, Chan, Azhar, Hui, Yuen, Coronaviruses-drug discovery and therapeutic options, Nat. Rev. Drug Discov,
doi:10.1038/nrd.2015.37DOI record:
{
"DOI": "10.1016/j.ijbiomac.2020.09.098",
"ISSN": [
"0141-8130"
],
"URL": "http://dx.doi.org/10.1016/j.ijbiomac.2020.09.098",
"alternative-id": [
"S0141813020344585"
],
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "International Journal of Biological Macromolecules"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.ijbiomac.2020.09.098"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2020 Elsevier B.V. All rights reserved."
}
],
"author": [
{
"affiliation": [],
"family": "Parvez",
"given": "Md. Sorwer Alam",
"sequence": "first"
},
{
"affiliation": [],
"family": "Karim",
"given": "Md. Adnan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hasan",
"given": "Mahmudul",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jaman",
"given": "Jomana",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Karim",
"given": "Ziaul",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tahsin",
"given": "Tohura",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hasan",
"given": "Md. Nazmul",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hosen",
"given": "Mohammad Jakir",
"sequence": "additional"
}
],
"container-title": "International Journal of Biological Macromolecules",
"container-title-short": "International Journal of Biological Macromolecules",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2020,
9,
17
]
],
"date-time": "2020-09-17T10:18:58Z",
"timestamp": 1600337938000
},
"deposited": {
"date-parts": [
[
2022,
6,
25
]
],
"date-time": "2022-06-25T20:56:02Z",
"timestamp": 1656190562000
},
"funder": [
{
"DOI": "10.13039/501100007944",
"doi-asserted-by": "publisher",
"name": "Shahjalal University of Science and Technology"
},
{
"name": "Bangladesh Bureau of Educational Information and Statistics"
},
{
"DOI": "10.13039/501100002701",
"doi-asserted-by": "publisher",
"name": "Ministry of Education"
}
],
"indexed": {
"date-parts": [
[
2024,
3,
16
]
],
"date-time": "2024-03-16T18:58:43Z",
"timestamp": 1710615523569
},
"is-referenced-by-count": 51,
"issued": {
"date-parts": [
[
2020,
11
]
]
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2020,
11,
1
]
],
"date-time": "2020-11-01T00:00:00Z",
"timestamp": 1604188800000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S0141813020344585?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S0141813020344585?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "1787-1797",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2020,
11
]
]
},
"published-print": {
"date-parts": [
[
2020,
11
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"DOI": "10.3855/jidc.12600",
"article-title": "Similarity in case fatality rates (CFR) of COVID-19/SARS-COV-2 in Italy and China",
"author": "Porcheddu",
"doi-asserted-by": "crossref",
"first-page": "125",
"journal-title": "J. Infect. Dev. Count.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0005",
"volume": "14",
"year": "2020"
},
{
"author": "Worldometer",
"first-page": "1",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0010",
"series-title": "Coronavirus Cases",
"year": "2020"
},
{
"author": "WHO",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0015",
"series-title": "Coronavirus Disease (COVID-19) Situation Dashboard",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)30154-9",
"article-title": "A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster",
"author": "Chan",
"doi-asserted-by": "crossref",
"journal-title": "Lancet",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0020",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)30211-7",
"article-title": "Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study",
"author": "Chen",
"doi-asserted-by": "crossref",
"journal-title": "Lancet",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0025",
"year": "2020"
},
{
"article-title": "Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China",
"author": "Yang",
"journal-title": "MedRxiv",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0030",
"year": "2020"
},
{
"article-title": "Caution on kidney dysfunctions of 2019-nCoV patients",
"author": "A.-2019-nCoV Volunteers",
"journal-title": "MedRxiv",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0035",
"year": "2020"
},
{
"article-title": "Severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2): an update",
"author": "Pal",
"journal-title": "Cureus",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0040",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1038/nrd.2015.37",
"article-title": "Coronaviruses—drug discovery and therapeutic options",
"author": "Zumla",
"doi-asserted-by": "crossref",
"first-page": "327",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0045",
"volume": "15",
"year": "2016"
},
{
"DOI": "10.1016/j.lfs.2020.117592",
"article-title": "Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study",
"author": "Elfiky",
"doi-asserted-by": "crossref",
"first-page": "117592",
"journal-title": "Life Sci.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0050",
"year": "2020"
},
{
"article-title": "The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase",
"author": "Lung",
"first-page": "jmv.25761",
"journal-title": "J. Med. Virol.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0055",
"year": "2020"
},
{
"DOI": "10.1038/d41573-020-00016-0",
"article-title": "Therapeutic options for the 2019 novel coronavirus (2019-nCoV)",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "149",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0060",
"volume": "19",
"year": "2020"
},
{
"DOI": "10.1016/j.antiviral.2013.09.015",
"article-title": "Favipiravir (T-705), a novel viral RNA polymerase inhibitor",
"author": "Furuta",
"doi-asserted-by": "crossref",
"first-page": "446",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0065",
"volume": "100",
"year": "2013"
},
{
"DOI": "10.1038/s41422-020-0282-0",
"article-title": "Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "269",
"journal-title": "Cell Res.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0070",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1128/mBio.00221-18",
"article-title": "Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease",
"author": "Agostini",
"doi-asserted-by": "crossref",
"journal-title": "MBio",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0075",
"volume": "9",
"year": "2018"
},
{
"author": "Zhang",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0080",
"series-title": "Binding Mechanism of Remdesivir to SARS-CoV-2 RNA Dependent RNA Polymerase",
"year": "2020"
},
{
"DOI": "10.1038/d41587-020-00003-1",
"article-title": "Coronavirus puts drug repurposing on the fast track",
"author": "Harrison",
"doi-asserted-by": "crossref",
"journal-title": "Nat. Biotechnol.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0085",
"year": "2020"
},
{
"DOI": "10.1038/sj.bjp.0707305",
"article-title": "In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling",
"author": "Ekins",
"doi-asserted-by": "crossref",
"first-page": "9",
"journal-title": "Br. J. Pharmacol.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0090",
"volume": "152",
"year": "2007"
},
{
"article-title": "Informatics and modeling challenges in fragment-based drug discovery",
"author": "Hubbard",
"first-page": "289",
"journal-title": "Curr. Opin. Drug Discov. Dev.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0095",
"volume": "10",
"year": "2007"
},
{
"DOI": "10.3389/fimmu.2019.00531",
"article-title": "Drug repurposing approaches for the treatment of influenza viral infection: reviving old drugs to fight against a long-lived enemy",
"author": "Pizzorno",
"doi-asserted-by": "crossref",
"journal-title": "Front. Immunol.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0100",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1016/j.tim.2018.04.004",
"article-title": "Drug repurposing for viral infectious diseases: how far are we?",
"author": "Mercorelli",
"doi-asserted-by": "crossref",
"first-page": "865",
"journal-title": "Trends Microbiol.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0105",
"volume": "26",
"year": "2018"
},
{
"author": "Li",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0110",
"series-title": "Network Bioinformatics Analysis Provides Insight Into Drug Repurposing for COVID-2019",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkv1290",
"article-title": "Database resources of the National Center for Biotechnology Information",
"author": "Resource Coordinators",
"doi-asserted-by": "crossref",
"first-page": "D7",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0115",
"volume": "44",
"year": "2016"
},
{
"article-title": "The RCSB protein data bank: integrative view of protein, gene and 3D structural information",
"author": "Rose",
"first-page": "D271",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0120",
"volume": "45",
"year": "2017"
},
{
"DOI": "10.1093/nar/gkx1037",
"article-title": "DrugBank 5.0: a major update to the DrugBank database for 2018",
"author": "Wishart",
"doi-asserted-by": "crossref",
"first-page": "D1074",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0125",
"volume": "46",
"year": "2018"
},
{
"article-title": "AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading",
"author": "Trott",
"journal-title": "J. Comput. Chem.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0130",
"volume": "31",
"year": "2009"
},
{
"article-title": "Pymol: an open-source molecular graphics tool",
"author": "DeLano",
"first-page": "82",
"issue": "1",
"journal-title": "CCP4 Newsl Prot. Crystallogr",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0135",
"volume": "40",
"year": "2002"
},
{
"DOI": "10.1021/ci200227u",
"article-title": "LigPlot+: multiple ligand-protein interaction diagrams for drug discovery",
"author": "Laskowski",
"doi-asserted-by": "crossref",
"first-page": "2778",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0140",
"volume": "51",
"year": "2011"
},
{
"author": "Biovia",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0145",
"series-title": "Discovery Studio Modeling Environment, Release 2017",
"year": "2016"
},
{
"DOI": "10.1093/nar/gkv315",
"article-title": "PLIP — fully automated protein-ligand interaction profiler",
"author": "Salentin",
"doi-asserted-by": "crossref",
"first-page": "W443",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0150",
"volume": "43",
"year": "2015"
},
{
"DOI": "10.1089/cmb.2007.0130",
"article-title": "Deterministic pharmacophore detection via multiple flexible alignment of drug-like molecules",
"author": "Schneidman-Duhovny",
"doi-asserted-by": "crossref",
"first-page": "737",
"journal-title": "J. Comput. Biol.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0155",
"volume": "15",
"year": "2008"
},
{
"DOI": "10.1093/nar/gkn187",
"article-title": "PharmaGist: a webserver for ligand-based pharmacophore detection",
"author": "Schneidman-Duhovny",
"doi-asserted-by": "crossref",
"first-page": "W223",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0160",
"volume": "36",
"year": "2008"
},
{
"DOI": "10.1093/nar/gks378",
"article-title": "ZINCPharmer: pharmacophore search of the ZINC database",
"author": "Koes",
"doi-asserted-by": "crossref",
"first-page": "W409",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0165",
"volume": "40",
"year": "2012"
},
{
"DOI": "10.1021/ci049714+",
"article-title": "ZINC — a free database of commercially available compounds for virtual screening",
"author": "Irwin",
"doi-asserted-by": "crossref",
"first-page": "177",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0170",
"volume": "45",
"year": "2005"
},
{
"DOI": "10.1038/srep42717",
"article-title": "SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules",
"author": "Daina",
"doi-asserted-by": "crossref",
"journal-title": "Sci. Rep.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0175",
"volume": "7",
"year": "2017"
},
{
"article-title": "Bioavailability prediction of phytochemicals present in Calotropis procera (Aiton) R. Br. by using Swiss-ADME tool",
"author": "Tripathi",
"first-page": "147",
"journal-title": "World Sci. News",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0180",
"volume": "131",
"year": "2019"
},
{
"article-title": "In vitro ADME studies of TUG-891, a GPR-120 inhibitor using SWISS ADME predictor",
"author": "Mishra",
"first-page": "366",
"journal-title": "J. Drug Deliv. Ther.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0185",
"volume": "9",
"year": "2019"
},
{
"key": "10.1016/j.ijbiomac.2020.09.098_bb0190",
"series-title": "Molecular Properties Prediction — Osiris Property Explorer",
"year": "2020"
},
{
"DOI": "10.1021/ci100104j",
"article-title": "Estimation of ADME properties with substructure pattern recognition",
"author": "Shen",
"doi-asserted-by": "crossref",
"first-page": "1034",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0195",
"volume": "50",
"year": "2010"
},
{
"DOI": "10.1021/ci300400a",
"article-title": "In silico prediction of chemical ames mutagenicity",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "2840",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0200",
"volume": "52",
"year": "2012"
},
{
"DOI": "10.1002/minf.201400127",
"article-title": "In silico estimation of chemical carcinogenicity with binary and ternary classification methods",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "228",
"journal-title": "Mol. Inform.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0205",
"volume": "34",
"year": "2015"
},
{
"author": "Schneidman-duhovny",
"first-page": "363",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0210",
"series-title": "PatchDock and SymmDock : Servers for Rigid and Symmetric Docking",
"volume": "vol. 33",
"year": "2005"
},
{
"author": "Hsu",
"first-page": "1",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0215",
"series-title": "iGEMDOCK: A Graphical Environment of Enhancing GEMDOCK Using Pharmacological Interactions and Post-screening Analysis",
"volume": "vol. 12",
"year": "2011"
},
{
"author": "Leach",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0220",
"series-title": "Molecular Modelling: Principles and Applications",
"year": "2001"
},
{
"author": "Yang",
"first-page": "1",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0225",
"series-title": "LARMD : Integration of Bioinformatic Resources to Profile Ligand-driven Protein Dynamics With a Case on the Activation of Estrogen Receptor",
"volume": "vol. 00",
"year": "2019"
},
{
"author": "Case",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0230",
"series-title": "The Amber Biomolecular Simulation Programs",
"volume": "12255",
"year": "2005"
},
{
"DOI": "10.1002/cmdc.201600182",
"article-title": "A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules",
"author": "Daina",
"doi-asserted-by": "crossref",
"first-page": "1117",
"journal-title": "ChemMedChem",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0235",
"volume": "11",
"year": "2016"
},
{
"DOI": "10.7861/clinmed.2019-coron",
"article-title": "What we know so far: COVID-19 current clinical knowledge and research",
"author": "Lake",
"doi-asserted-by": "crossref",
"first-page": "124",
"journal-title": "Clin. Med.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0240",
"volume": "20",
"year": "2020"
},
{
"DOI": "10.1186/s13578-020-00404-4",
"article-title": "SARS-CoV-2 and COVID-19: the most important research questions",
"author": "Yuen",
"doi-asserted-by": "crossref",
"first-page": "40",
"journal-title": "Cell Biosci",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0245",
"volume": "10",
"year": "2020"
},
{
"article-title": "Sensitivity of chest CT for COVID-19: comparison to RT-PCR",
"author": "Fang",
"first-page": "200432",
"journal-title": "Radiology.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0250",
"year": "2020"
},
{
"DOI": "10.5582/ddt.2020.01012",
"article-title": "Discovering drugs to treat coronavirus disease 2019 (COVID-19)",
"author": "Dong",
"doi-asserted-by": "crossref",
"first-page": "58",
"journal-title": "Drug Discov. Ther.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0255",
"volume": "14",
"year": "2020"
},
{
"article-title": "COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression",
"author": "Zhou",
"journal-title": "J. Antimicrob. Chemother.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0260",
"year": "2020"
},
{
"DOI": "10.1038/nrd2424",
"article-title": "The design of drugs for HIV and HCV",
"author": "De Clercq",
"doi-asserted-by": "crossref",
"first-page": "1001",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0265",
"volume": "6",
"year": "2007"
},
{
"article-title": "Purification of Zika virus RNA-dependent RNA polymerase and its use to identify small-molecule Zika inhibitors",
"author": "Xu",
"first-page": "727",
"journal-title": "J. Antimicrob. Chemother.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0270",
"volume": "72",
"year": "2017"
},
{
"article-title": "RNA-dependent RNA polymerases and their emerging roles in antiviral therapy",
"author": "Ganeshpurkar",
"first-page": "1",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0275",
"series-title": "Viral Polym.",
"year": "2019"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"article-title": "The FDA-approved drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro",
"author": "Caly",
"doi-asserted-by": "crossref",
"first-page": "104787",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0280",
"year": "2020"
},
{
"DOI": "10.1093/clinids/11.Supplement_7.S1648",
"article-title": "Antiviral chemotherapy for infection with human immunodeficiency virus",
"author": "Polsky",
"doi-asserted-by": "crossref",
"first-page": "S1648",
"journal-title": "Rev. Infect. Dis.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0285",
"volume": "11",
"year": "1989"
},
{
"DOI": "10.2174/1568005053174627",
"article-title": "Orthopoxvirus targets for the development of antiviral therapies",
"author": "Prichard",
"doi-asserted-by": "crossref",
"first-page": "17",
"journal-title": "Curr. Drug Targets. Infect. Disord.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0290",
"volume": "5",
"year": "2005"
},
{
"author": "Agrawal",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0295",
"series-title": "Benchmarking of Different Molecular Docking Methods for Protein-Peptide Docking",
"volume": "vol. 19",
"year": "2019"
},
{
"DOI": "10.2142/biophysico.13.0_181",
"article-title": "Molecular dynamics analysis to evaluate docking pose prediction",
"author": "Sakano",
"doi-asserted-by": "crossref",
"first-page": "181",
"journal-title": "Biophys. Physicobiol.",
"key": "10.1016/j.ijbiomac.2020.09.098_bb0300",
"volume": "13",
"year": "2016"
}
],
"reference-count": 60,
"references-count": 60,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S0141813020344585"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Molecular Biology",
"General Medicine",
"Biochemistry",
"Structural Biology"
],
"subtitle": [],
"title": "Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1016/elsevier_cm_policy",
"volume": "163"
}