A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients
Ph.D Dharmendra Kumar Maurya
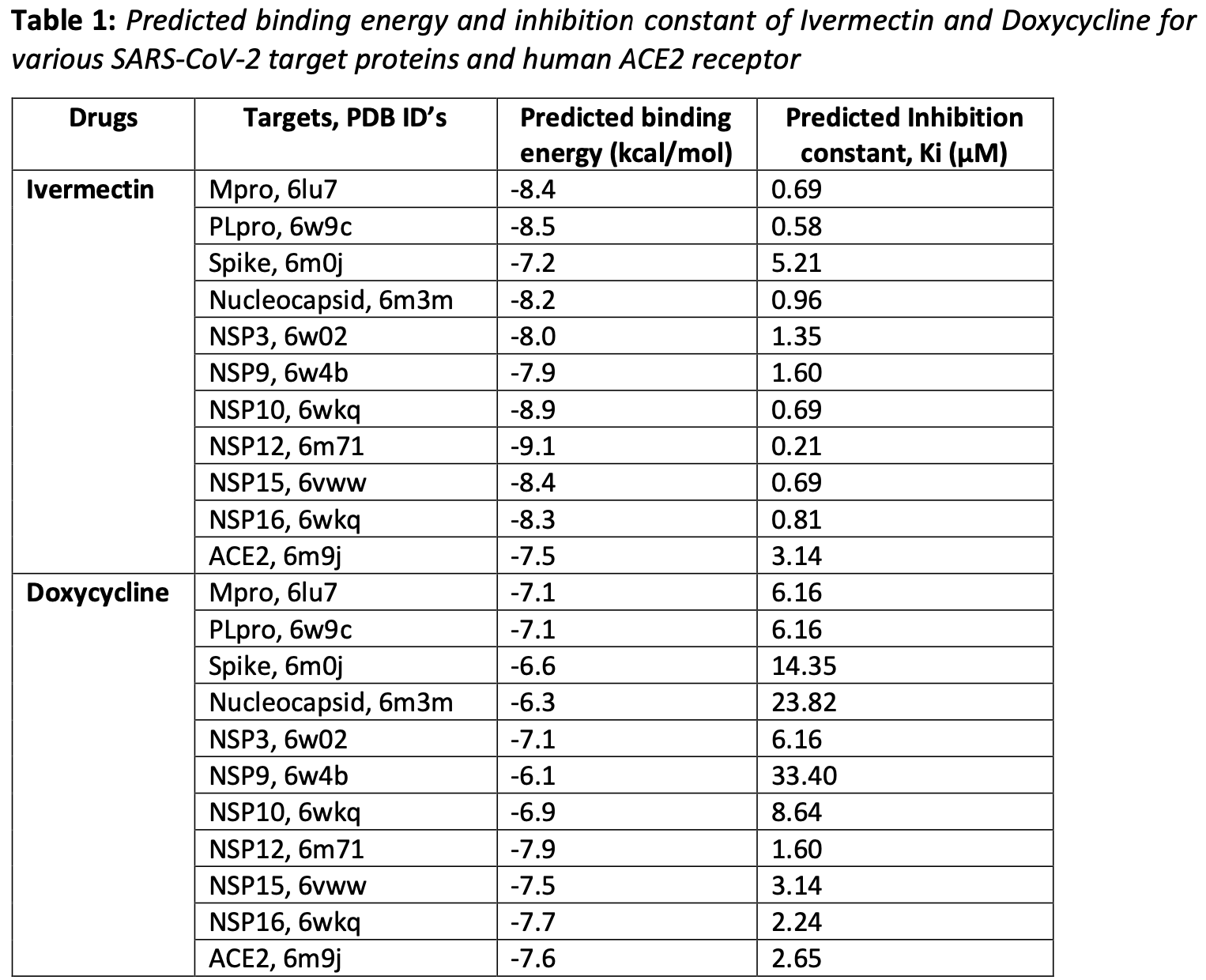
The current outbreak of the corona virus disease 2019 , has affected almost entire world and become pandemic now. Currently, there is neither any FDA approved drugs nor any vaccines available to control it. Very recently in Bangladesh, a group of doctors reported astounding success in treating patients suffering from COVID-19 with two commonly used drugs, Ivermectin and Doxycycline. In the current study we have explored the possible mechanism by which these drugs might have worked for the positive response in the COVID-19 patients. To explore the mechanism we have used molecular docking and molecular dynamics simulation approach. Effectiveness of Ivermectin and doxycycline were evaluated against Main Protease (Mpro), Spike (S) protein, Nucleocapsid (N), RNA-dependent RNA polymerase (RdRp, NSP12), ADP Ribose Phosphatase (NSP3), Endoribonuclease (NSP15) and methyltransferase (NSP10-NSP16 complex) of SARS-CoV-2 as well as human angiotensin converting enzyme 2 (ACE2) receptor. Our study shows that both Ivermectin and doxycycline have significantly bind with SARS-CoV-2 proteins but Ivermectin was better binding than doxycycline. Ivermectin showed a perfect binding site to the Spike-RBD and ACE2 interacting region indicating that it might be interfering in the interaction of spike with ACE2 and preventing the viral entry in to the host cells. Ivermectin also exhibited significant binding affinity with different SARS-CoV-2 structural and non-structural proteins (NSPs) which have diverse functions in virus life cycle. Significant binding of Ivermectin with RdRp indicate its role in the inhibition of the viral replication and ultimately impeding the multiplication of the virus. Ivermectin also possess significant binding affinity with NSP3, NSP10, NSP15 and NSP16 which helps virus in escaping from host immune system. Molecular dynamics simulation study shows that binding of the Ivermectin with Mpro, Spike, NSP3, NSP16 and ACE2 was quiet stable. Thus, our docking and simulation studies reveal that combination of Ivermectin and doxycycline might be executing the effect by inhibition of viral entry and enhance viral load clearance by targeting various viral functional proteins.
Declaration of Conflicting Interests The author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
Abraham, Murtola, Schulz, Páll, Smith et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX,
doi:10.1016/j.softx.2015.06.001Aggarwal, Jain, Talapatra, Yadav, Gupta et al., Evaluation of role of doxycycline (a matrix metalloproteinase inhibitor) on renal functions in patients of diabetic nephropathy, Ren Fail,
doi:10.3109/0886022x.2010.502606Bhardwaj, Liu, Leibowitz, Kao, The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein, Journal of virology,
doi:10.1128/JVI.07012-11Buonfrate, Salas-Coronas, Muñoz, Maruri, Rodari et al., Multiple-dose versus single-dose ivermectin for Strongyloides stercoralis infection (Strong Treat 1 to 4): a multicentre, open-label, phase 3, randomised controlled superiority trial, The Lancet Infectious Diseases,
doi:10.1016/S1473-3099(19)30289-0Egloff, Ferron, Campanacci, Longhi, Rancurel et al., The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world, Proceedings of the National Academy of Sciences of the United States of America,
doi:10.1073/pnas.0307877101González Canga, Sahagún Prieto, Diez Liébana, Fernández Martínez, Sierra et al., The Pharmacokinetics and Interactions of Ivermectin in Humans-A Mini-review, The AAPS Journal,
doi:10.1208/s12248-007-9000-9Götz, Magar, Dornfeld, Giese, Pohlmann et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific reports,
doi:10.1038/srep23138He, Marneros, Doxycycline inhibits polarization of macrophages to the proangiogenic M2-type and subsequent neovascularization, The Journal of biological chemistry,
doi:10.1074/jbc.m113.535765Henehan, Montuno, De Benedetto, Doxycycline as an anti-inflammatory agent: updates in dermatology, J Eur Acad Dermatol Venereol,
doi:10.1111/jdv.14345Hu, Chen, Li, Dou, Zhou et al., Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9, Protein Sci,
doi:10.1002/pro.3150Ivanov, Hertzig, Rozanov, Bayer, Thiel et al., Major genetic marker of nidoviruses encodes a replicative endoribonuclease, Proc Natl Acad Sci U S A,
doi:10.1073/pnas.0403127101Kang, Bhardwaj, Li, Palaninathan, Sacchettini et al., Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease, J Virol,
doi:10.1128/JVI.00547-07Lundberg, Pinkham, Baer, Amaya, Narayanan et al., Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication, Antiviral Res,
doi:10.1016/j.antiviral.2013.10.004Maurya, Sharma, Evaluation of Traditional Ayurvedic Preparation for Prevention and Management of the Novel Coronavirus (SARS-CoV-2) Using Molecular Docking Approach, ChemRxiv (Preprints,
doi:10.26434/chemrxiv.12110214Mcbride, Van Zyl, Fielding, The coronavirus nucleocapsid is a multifunctional protein, Viruses,
doi:10.3390/v6082991Mielech, Chen, Mesecar, Baker, Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities, Virus Res,
doi:10.1016/j.virusres.2014.01.025Pettersen, Goddard, Huang, Couch, Greenblatt et al., UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem,
doi:10.1002/jcc.20084Phillips, Gallagher, Weiss, Neurovirulent Murine Coronavirus JHM.SD Uses Cellular Zinc Metalloproteases for Virus Entry and Cell-Cell Fusion, Journal of Virology,
doi:10.1128/JVI.01564-16Posthuma, Nedialkova, Zevenhoven-Dobbe, Blokhuis, Gorbalenya et al., Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle, J Virol,
doi:10.1128/JVI.80.4.1653-1661.2006Prajapat, Sarma, Shekhar, Avti, Sinha et al., Drug targets for corona virus: A systematic review, Indian journal of pharmacology,
doi:10.4103/ijpSutton, Fry, Carter, Sainsbury, Walter et al., The nsp9 replicase protein of SARS-coronavirus, structure and functional insights, Structure,
doi:10.1016/j.str.2004.01.016Tay, Fraser, Chan, Moreland, Rathore et al., Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin, Antiviral Res,
doi:10.1016/j.antiviral.2013.06.002Van Aalten, Bywater, Findlay, Hendlich, Hooft et al., PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules, Journal of Computer-Aided Molecular Design,
doi:10.1007/BF00355047Wu, Liu, Yang, Zhang, Zhong et al., Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods, Acta Pharmaceutica Sinica B,
doi:10.1016/j.apsb.2020.02.008Yang, Atkinson, Wang, Lee, Bogoyevitch et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Res,
doi:10.1016/j.antiviral.2020.104760Yang, Chen, Tu, Yen, Yang, Combinatorial Computational Approaches to Identify Tetracycline Derivatives as Flavivirus Inhibitors, PLOS ONE,
doi:10.1371/journal.pone.0000428Zhang, Penninger, Li, Zhong, Slutsky, Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target, Intensive Care Medicine,
doi:10.1007/s00134-020-05985-9Zhou, Hou, Shen, Huang, Martin et al., Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2, Cell Discovery,
doi:10.1038/s41421-020-0153-3DOI record:
{
"DOI": "10.26434/chemrxiv.12630539.v1",
"URL": "http://dx.doi.org/10.26434/chemrxiv.12630539.v1",
"abstract": "<jats:p>The current outbreak of the corona virus disease 2019 (COVID-19), has affected almost entire world and become pandemic now. Currently, there is neither any FDA approved drugs nor any vaccines available to control it. Very recently in Bangladesh, a group of doctors reported astounding success in treating patients suffering from COVID-19 with two commonly used drugs, Ivermectin and Doxycycline. In the current study we have explored the possible mechanism by which these drugs might have worked for the positive response in the COVID-19 patients. To explore the mechanism we have used molecular docking and molecular dynamics simulation approach. Effectiveness of Ivermectin and doxycycline were evaluated against Main Protease (Mpro), Spike (S) protein, Nucleocapsid (N), RNA-dependent RNA polymerase (RdRp, NSP12), ADP Ribose Phosphatase (NSP3), Endoribonuclease (NSP15) and methyltransferase (NSP10-NSP16 complex) of SARS-CoV-2 as well as human angiotensin converting enzyme 2 (ACE2) receptor. Our study shows that both Ivermectin and doxycycline have significantly bind with SARS-CoV-2 proteins but Ivermectin was better binding than doxycycline. Ivermectin showed a perfect binding site to the Spike-RBD and ACE2 interacting region indicating that it might be interfering in the interaction of spike with ACE2 and preventing the viral entry in to the host cells. Ivermectin also exhibited significant binding affinity with different SARS-CoV-2 structural and non-structural proteins (NSPs) which have diverse functions in virus life cycle. Significant binding of Ivermectin with RdRp indicate its role in the inhibition of the viral replication and ultimately impeding the multiplication of the virus. Ivermectin also possess significant binding affinity with NSP3, NSP10, NSP15 and NSP16 which helps virus in escaping from host immune system. Molecular dynamics simulation study shows that binding of the Ivermectin with Mpro, Spike, NSP3, NSP16 and ACE2 was quiet stable. Thus, our docking and simulation studies reveal that combination of Ivermectin and doxycycline might be executing the effect by inhibition of viral entry and enhance viral load clearance by targeting various viral functional proteins.</jats:p>",
"accepted": {
"date-parts": [
[
2020,
7,
9
]
]
},
"author": [
{
"ORCID": "http://orcid.org/0000-0003-0909-363X",
"affiliation": [
{
"name": "Radiation Biology & Health Sciences Division, Bhabha Atomic Research Centre, Mumbai"
}
],
"authenticated-orcid": false,
"family": "Maurya",
"given": "Dharmendra Kumar",
"sequence": "first"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2020,
7,
9
]
],
"date-time": "2020-07-09T13:22:49Z",
"timestamp": 1594300969000
},
"deposited": {
"date-parts": [
[
2021,
10,
21
]
],
"date-time": "2021-10-21T20:59:35Z",
"timestamp": 1634849975000
},
"group-title": "Chemistry",
"indexed": {
"date-parts": [
[
2024,
2,
5
]
],
"date-time": "2024-02-05T05:24:59Z",
"timestamp": 1707110699874
},
"is-referenced-by-count": 11,
"issued": {
"date-parts": [
[
2020,
7,
9
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0/",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2020,
7,
9
]
],
"date-time": "2020-07-09T00:00:00Z",
"timestamp": 1594252800000
}
}
],
"link": [
{
"URL": "https://chemrxiv.org/engage/api-gateway/chemrxiv/assets/orp/resource/item/60c74d85842e655304db34b6/original/a-combination-of-ivermectin-and-doxycycline-possibly-blocks-the-viral-entry-and-modulate-the-innate-immune-response-in-covid-19-patients.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "316",
"original-title": [],
"posted": {
"date-parts": [
[
2020,
7,
9
]
]
},
"prefix": "10.26434",
"published": {
"date-parts": [
[
2020,
7,
9
]
]
},
"publisher": "American Chemical Society (ACS)",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://chemrxiv.org/engage/chemrxiv/article-details/60c74d85842e655304db34b6"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "A Combination of Ivermectin and Doxycycline Possibly Blocks the Viral Entry and Modulate the Innate Immune Response in COVID-19 Patients",
"type": "posted-content"
}