Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches
Hourieh Kalhor, Solmaz Sadeghi, Hoda Abolhasani, Reyhaneh Kalhor, Hamzeh Rahimi
Journal of Biomolecular Structure and Dynamics, doi:10.1080/07391102.2020.1824816
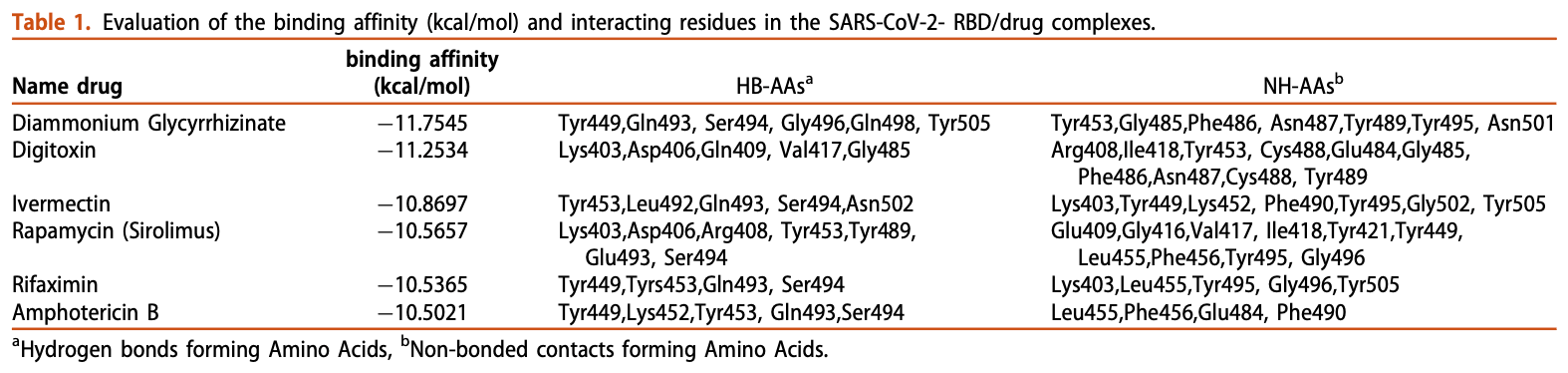
Most recently, the new coronavirus (SARS-CoV-2) has been recognized as a pandemic by the World Health Organization (WHO) while this virus shares substantial similarity with SARS-CoV. So far, no definitive vaccine or drug has been developed to cure Covid-19 disease, since many important aspects about Covid-19 such as pathogenesis and proliferation pathways are still unclear. It was proven that human ACE2 is the main receptor for the entry of Covid-19 into lower respiratory tract epithelial cells through interaction with SARS-CoV-2 S protein. Based on this observation, it is expected that the virus infection can be inhibited if protein-protein interaction is prevented. In this study, using structurebased virtual screening of FDA databases, several lead drugs were discovered based on the ACE2binding pocket of SARS-CoV-2 S protein. Then, binding affinity, binding modes, critical interactions, and pharmaceutical properties of the lead drugs were evaluated. Among the previously approved drugs, Diammonium Glycyrrhizinate, Digitoxin, Ivermectin, Rapamycin, Rifaximin, and Amphotericin B represented the most desirable features, and can be possible candidates for Covid-19 therapies. Furthermore, molecular dynamics (MD) simulation was accomplished for three S protein/drug complexes with the highest binding affinity and best conformation and binding free energies were also computed with the Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) method. Results demonstrated the stable binding of these compounds to the S protein; however, in order to confirm the curative effect of these drugs, clinical trials must be done.
References
Abraham, Murtola, Schulz, Smith, Hess et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX,
doi:10.1016/j.softx.2015.06.001Agostini, Andres, Sims, Graham, Sheahan et al., Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease, mBio,
doi:10.1128/mBio.00221-18Alobid, Bernal, Calvo, Vilaseca, Berenguer et al., Treatment of rhinocerebral mucormycosis by combination of endoscopic sinus debridement and amphotericin B, American Journal of Rhinology,
doi:10.1177/1945892401Amarelle, Lecuona, The antiviral effects of Na, K-ATPase inhibition: A minireview, International Journal of Molecular Sciences,
doi:10.3390/ijms19082154Athanasios, Psychos, Domenikos, Communication: Novel drug combination Doxycycline-Melatonin-Digoxin (D-M-D) for possible Covid-19 treatment, Journal of Modern Medicinal Chemistry,
doi:10.12970/2308-8044.2020.08.01Bussi, Donadio, Parrinello, Canonical sampling through velocity rescaling, The Journal of Chemical Physics,
doi:10.1063/1.2408420Chang, Tung, Lee, Chen, Hsiao et al., Potential Therapeutic Agents for COVID-19 Based on the Analysis of Protease and RNA Polymerase Docking, Preprints,
doi:10.20944/preprints202002.0242.v2Cinatl, Morgenstern, Bauer, Chandra, Rabenau et al., Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus, The Lancet,
doi:10.1016/S0140-6736(03)13615-XDarden, York, Pedersen, Particle mesh Ewald: An NÁ log (N) method for Ewald sums in large systems, The Journal of Chemical Physics,
doi:10.1063/1.464397De Oliveira, Rocha, Paluch, Costa, Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening, Journal of Biomolecular Structure and Dynamics,
doi:10.1080/07391102.2020.1772885Delano, Pymol: An open-source molecular graphics tool, CCP4 Newsletter on Protein Crystallography
Elfiky, Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study, Life Sciences,
doi:10.1016/j.lfs.2020.117592Feng, Wang, Yao, Zhang, Tian, Diammonium glycyrrhizinate, a component of traditional Chinese medicine Gan-Cao, prevents murine T-cell-mediated fulminant hepatitis in IL-10-and IL-6dependent manners, International Immunopharmacology,
doi:10.1016/j.intimp.2007.05.011Gupta, Ammonium glycyrrhizinate: a compherensive review of its traditionaluse, phytochemistry, pharmacology & safety, Asian Journal of Pharmaceutical Education and Research
G€ Otz, Magar, Dornfeld, Giese, Pohlmann et al., Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import, Scientific Reports,
doi:10.1038/srep23138Hackett, Sylvester, Joss, Calvin, Synergistic effect of rifamycin derivatives and amphotericin B on viral transformation of a murine cell line, Proceedings of the National Academy of Sciences,
doi:10.1073/pnas.69.12.3653Hess, Bekker, Berendsen, Fraaije, Ito et al., Mechanism of inhibitory effect of glycyrrhizin on replication of human immunodeficiency virus (HIV), Journal of Computational Chemistry,
doi:10.1016/0166-3542(88)90047-2Kalhor, Rahimi, Akbari Eidgahi, Teimoori-Toolabi, Novel small molecules against two binding sites of Wnt2 protein as potential drug candidates for colorectal cancer: A structure based virtual screening approach, Iranian Journal of Pharmaceutical Research,
doi:10.22037/IJPR.2019.15297.13037Kalhor, Sadeghi, Marashiyan, Kalhor, Aghaei Gharehbolagh et al., Identification of new DNA gyrase inhibitors based on bioactive compounds from streptomyces: Structure-based virtual screening and molecular dynamics simulations approaches, Journal of Biomolecular Structure & Dynamics,
doi:10.1080/07391102.2019.1588784Kandeel, Al-Nazawi, Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease, Life Sciences,
doi:10.1016/j.lfs.2020.117627Koes, Baumgartner, Camacho, Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise, Journal of Chemical Information and Modeling,
doi:10.1021/ci300604zKumari, Kumar, Consortium, Lynn, g_mmpbsaA A GROMACS tool for high-throughput MM-PBSA calculations, Journal of Chemical Information and Modeling,
doi:10.1021/ci500020mLaskowski, Swindells, LigPlotþ: Multiple ligand-protein interaction diagrams for drug discovery, ACS Publications,
doi:10.1021/ci200227uLi, Li, Farzan, Harrison, Structure of SARS coronavirus spike receptor-binding domain complexed with receptor, Science,
doi:10.1126/science.1116480Lipinski, Lombardo, Dominy, Feeney, Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings, Advanced Drug Delivery Reviews,
doi:10.1016/S0169-409X(00)00129-0Lundberg, Pinkham, Baer, Amaya, Narayanan et al., Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication, Antiviral Research,
doi:10.1016/j.antiviral.2013.10.004Mohammadi, Kometiani, Xie, Askari, Role of protein kinase C in the signal pathways that link Naþ/Kþ-ATPase to ERK1/2, Journal of Biological Chemistry,
doi:10.1074/jbc.M107892200Morgenstern, Michaelis, Baer, Doerr, Cinatl, Ribavirin and interferon-b synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines, Biochemical and Biophysical Research Communications,
doi:10.1016/j.bbrc.2004.11.128Morris, Huey, Lindstrom, Sanner, Belew et al., AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, Journal of Computational Chemistry,
doi:10.1002/jcc.21256Mukhtar, Mukhtar, Coronavirus (COVID-19): Let's prevent not panic, Journal of Ayub Medical College Abbottabad
Numazaki, Nagata, Sato, Chiba, Effect of glycyrrhizin, cyclosporin A, and tumor necrosis factor a on infection of U-937 and MRC-5 cells by human cytomegalovirus, Journal of Leukocyte Biology,
doi:10.1002/jlb.55.1.24O'boyle, Banck, James, Morley, Vandermeersch et al., Open Babel: An open chemical toolbox, Journal of Cheminformatics,
doi:10.1021/ci200227uParrinello, Rahman, Polymorphic transitions in single crystals: A new molecular dynamics method Polymorphic transitions in single crystals: A new molecular dynamics method, Journal of Applied Physics,
doi:10.1063/1.328693Pollard, Pollard, Pollard, Classical drug digitoxin inhibits influenza cytokine storm, with implications for COVID-19 therapy, bioRxiv,
doi:10.1101/2020.04.09.034983Rismanbaf, Potential treatments for COVID-19; a narrative literature review, Archives of Academic Emergency Medicine
Sander, Freyss, Von Korff, Rufener, DataWarrior: An open-source program for chemistry aware data visualization and analysis, Journal of Chemical Information and Modeling,
doi:10.1021/ci500588jSch€ Uttelkopf, Van Aalten, PRODRG: A tool for highthroughput crystallography of protein-ligand complexes, Acta Crystallographica Section D Biological Crystallography,
doi:10.1107/S0907444904011679Shanmugaraj, Malla, Phoolcharoen, Emergence of novel coronavirus 2019-nCoV: Need for rapid vaccine and biologics development, Pathogens,
doi:10.3390/pathogens9020148Shiri, Pirhadi, Ghasemi, Dynamic structure based pharmacophore modeling of the Acetylcholinesterase reveals several potential inhibitors, Journal of Biomolecular Structure & Dynamics,
doi:10.1080/07391102.2018.1468281Shiri, Pirhadi, Rahmani, Identification of new potential HIV-1 reverse transcriptase inhibitors by QSAR modeling and structure-based virtual screening, Journal of Receptor and Signal Transduction Research,
doi:10.1080/10799893.2017.1414844Song, Gui, Wang, Xiang, Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2, PLOS Pathogens,
doi:10.1371/journal.ppat.1007236Tay, Fraser, Chan, Moreland, Rathore et al., Nuclear ocalization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENVserotypes by the inhibitor Ivermectin, Antiviral Research,
doi:10.1016/j.antiviral.2013.06.002Thakur, Raj, Pharmacological perspective of Glycyrrhiza glabra Linn: A mini-review, Journal of Analytical & Pharmaceutical Research,
doi:10.15406/japlr.2017.05.00156Utsunomiya, Kobayashi, Herndon, Pollard, Suzuki et al., Glycyrrhizin(20b-carboxy-11-oxo-30-norolean-12-en-3b-yl-2-Ob-d-glucopyranuronosyl-a-d-glucopyranosiduronic acid) improves the resistance of thermally injured mice to opportunistic infection of herpes simplex virus type 1, Antimicrobial Agents and Chemotherapy,
doi:10.1128/AAC.41.3.551Van Tilbeurgh, Bezzine, Cambillau, Verger, Carriere, Colipase: Structure and interaction with pancreatic lipase, Biochimica et Biophysica Acta (Bba) -Molecular and Cell Biology of Lipids,
doi:10.1016/S1388-1981(99)00149-3Visualizer, Version 4.5 (software
Wagstaff, Sivakumaran, Heaton, Harrich, Jans, Ivermectin is a specific inhibitor of importin a/b-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, The Biochemical Journal,
doi:10.1042/BJ20120150Wrapp, Wang, Corbett, Goldsmith, Hsieh et al., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science,
doi:10.1126/science.abb2507Xu, Chen, Wang, Feng, Zhou et al., Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission, Science China Life Sciences,
doi:10.1007/s11427-020-1637-5Zhou, Hou, Shen, Huang, Martin et al., Network-based drug repurposing for novel coronavirus 2019-nCoV/ SARS-CoV-2, Cell Discovery,
doi:10.1038/s41421-020-0153-3Zhu, Zhang, Wang, Li, Yang et al., A novel coronavirus from patients with pneumonia in China, 2019, The New England Journal of Medicine,
doi:10.1056/NEJMoa2001017DOI record:
{
"DOI": "10.1080/07391102.2020.1824816",
"ISSN": [
"0739-1102",
"1538-0254"
],
"URL": "http://dx.doi.org/10.1080/07391102.2020.1824816",
"alternative-id": [
"10.1080/07391102.2020.1824816"
],
"assertion": [
{
"label": "Peer Review Statement",
"name": "peerreview_statement",
"order": 1,
"value": "The publishing and review policy for this title is described in its Aims & Scope."
},
{
"URL": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=tbsd20",
"label": "Aim & Scope",
"name": "aims_and_scope_url",
"order": 2,
"value": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=tbsd20"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2020-08-01"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2020-09-12"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2020-09-24"
}
],
"author": [
{
"affiliation": [
{
"name": "Cellular and Molecular Research Center, Qom University of Medical Sciences, Qom, Iran"
},
{
"name": "Molecular Medicine Department, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran"
}
],
"family": "Kalhor",
"given": "Hourieh",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0001-5861-2606",
"affiliation": [
{
"name": "Department of Medical Biotechnology, School of Advanced Technologies in Medicine, Tehran University of Medical Sciences, Tehran, Iran"
}
],
"authenticated-orcid": false,
"family": "Sadeghi",
"given": "Solmaz",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9174-5304",
"affiliation": [
{
"name": "Cellular and Molecular Research Center, Qom University of Medical Sciences, Qom, Iran"
},
{
"name": "Spiritual Health Research Center, Qom University of Medical Sciences, Qom, Iran"
},
{
"name": "Department of Pharmacology, School of Medicine, Qom University of Medical Sciences, Qom, Iran"
}
],
"authenticated-orcid": false,
"family": "Abolhasani",
"given": "Hoda",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Cellular and Molecular Research Center, Qom University of Medical Sciences, Qom, Iran"
},
{
"name": "Department of Genetics, Colleague of Sciences, Kazerun branch, Islamic Azad University, Kazerun, Iran"
}
],
"family": "Kalhor",
"given": "Reyhaneh",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Molecular Medicine Department, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran"
}
],
"family": "Rahimi",
"given": "Hamzeh",
"sequence": "additional"
}
],
"container-title": "Journal of Biomolecular Structure and Dynamics",
"container-title-short": "Journal of Biomolecular Structure and Dynamics",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.tandfonline.com"
]
},
"created": {
"date-parts": [
[
2020,
9,
24
]
],
"date-time": "2020-09-24T12:10:25Z",
"timestamp": 1600949425000
},
"deposited": {
"date-parts": [
[
2022,
1,
31
]
],
"date-time": "2022-01-31T13:12:23Z",
"timestamp": 1643634743000
},
"indexed": {
"date-parts": [
[
2024,
2,
20
]
],
"date-time": "2024-02-20T10:18:49Z",
"timestamp": 1708424329552
},
"is-referenced-by-count": 30,
"issue": "3",
"issued": {
"date-parts": [
[
2020,
9,
24
]
]
},
"journal-issue": {
"issue": "3",
"published-print": {
"date-parts": [
[
2022,
2,
11
]
]
}
},
"language": "en",
"link": [
{
"URL": "https://www.tandfonline.com/doi/pdf/10.1080/07391102.2020.1824816",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "301",
"original-title": [],
"page": "1299-1315",
"prefix": "10.1080",
"published": {
"date-parts": [
[
2020,
9,
24
]
]
},
"published-online": {
"date-parts": [
[
2020,
9,
24
]
]
},
"published-print": {
"date-parts": [
[
2022,
2,
11
]
]
},
"publisher": "Informa UK Limited",
"reference": [
{
"DOI": "10.1016/j.softx.2015.06.001",
"doi-asserted-by": "publisher",
"key": "CIT0001"
},
{
"DOI": "10.1128/mBio.00221-18",
"doi-asserted-by": "publisher",
"key": "CIT0002"
},
{
"DOI": "10.1177/194589240101500508",
"doi-asserted-by": "publisher",
"key": "CIT0003"
},
{
"DOI": "10.3390/ijms19082154",
"doi-asserted-by": "publisher",
"key": "CIT0004"
},
{
"DOI": "10.1002/(SICI)1097-0142(19970415)79:8<1494::AID-CNCR8>3.0.CO;2-B",
"doi-asserted-by": "publisher",
"key": "CIT0005"
},
{
"DOI": "10.12970/2308-8044.2020.08.01",
"doi-asserted-by": "publisher",
"key": "CIT0006"
},
{
"DOI": "10.1016/0166-3542(87)90025-8",
"doi-asserted-by": "publisher",
"key": "CIT0007"
},
{
"DOI": "10.1063/1.2408420",
"doi-asserted-by": "publisher",
"key": "CIT0008"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"doi-asserted-by": "publisher",
"key": "CIT0009"
},
{
"DOI": "10.20944/preprints202002.0242.v2",
"doi-asserted-by": "publisher",
"key": "CIT0010"
},
{
"DOI": "10.1016/S0140-6736(03)13615-X",
"doi-asserted-by": "publisher",
"key": "CIT0011"
},
{
"DOI": "10.1063/1.464397",
"doi-asserted-by": "publisher",
"key": "CIT0012"
},
{
"DOI": "10.1080/07391102.2020.1772885",
"doi-asserted-by": "publisher",
"key": "CIT0013"
},
{
"author": "DeLano W. L.",
"first-page": "82",
"journal-title": "CCP4 Newsletter on Protein Crystallography",
"key": "CIT0014",
"volume": "40",
"year": "2002"
},
{
"DOI": "10.1016/j.lfs.2020.117477",
"doi-asserted-by": "publisher",
"key": "CIT0015"
},
{
"DOI": "10.1016/j.lfs.2020.117592",
"doi-asserted-by": "publisher",
"key": "CIT0016"
},
{
"DOI": "10.1016/j.intimp.2007.05.011",
"doi-asserted-by": "publisher",
"key": "CIT0017"
},
{
"DOI": "10.1038/srep23138",
"doi-asserted-by": "publisher",
"key": "CIT0018"
},
{
"DOI": "10.1073/pnas.69.12.3653",
"doi-asserted-by": "publisher",
"key": "CIT0019"
},
{
"DOI": "10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H",
"doi-asserted-by": "publisher",
"key": "CIT0020"
},
{
"DOI": "10.1016/0166-3542(88)90047-2",
"doi-asserted-by": "publisher",
"key": "CIT0021"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"doi-asserted-by": "publisher",
"key": "CIT0022"
},
{
"author": "Kalhor H.",
"issue": "2",
"journal-title": "Iranian Journal of Pharmaceutical Research",
"key": "CIT0023",
"volume": "19",
"year": "2020"
},
{
"DOI": "10.1080/07391102.2019.1588784",
"doi-asserted-by": "publisher",
"key": "CIT0024"
},
{
"DOI": "10.1016/j.lfs.2020.117627",
"doi-asserted-by": "publisher",
"key": "CIT0025"
},
{
"DOI": "10.1021/ci300604z",
"doi-asserted-by": "publisher",
"key": "CIT0026"
},
{
"DOI": "10.1021/ci500020m",
"doi-asserted-by": "publisher",
"key": "CIT0027"
},
{
"DOI": "10.1093/nar/gkn860",
"doi-asserted-by": "publisher",
"key": "CIT0028"
},
{
"author": "Laskowski R. A.",
"key": "CIT0029",
"volume-title": "LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery",
"year": "2011"
},
{
"DOI": "10.1016/j.cmet.2014.01.001",
"doi-asserted-by": "publisher",
"key": "CIT0030"
},
{
"DOI": "10.1126/science.1116480",
"doi-asserted-by": "publisher",
"key": "CIT0031"
},
{
"DOI": "10.1016/S0166-3542(03)00030-5",
"doi-asserted-by": "publisher",
"key": "CIT0032"
},
{
"DOI": "10.1016/j.addr.2012.09.019",
"doi-asserted-by": "publisher",
"key": "CIT0033"
},
{
"DOI": "10.1016/j.antiviral.2013.10.004",
"doi-asserted-by": "publisher",
"key": "CIT0034"
},
{
"DOI": "10.1074/jbc.M107892200",
"doi-asserted-by": "publisher",
"key": "CIT0035"
},
{
"DOI": "10.1016/j.bbrc.2004.11.128",
"doi-asserted-by": "publisher",
"key": "CIT0036"
},
{
"DOI": "10.1002/jcc.21256",
"doi-asserted-by": "publisher",
"key": "CIT0037"
},
{
"author": "Mukhtar F.",
"first-page": "141",
"issue": "1",
"journal-title": "Journal of Ayub Medical College Abbottabad",
"key": "CIT0038",
"volume": "32",
"year": "2020"
},
{
"DOI": "10.1002/jlb.55.1.24",
"doi-asserted-by": "publisher",
"key": "CIT0039"
},
{
"DOI": "10.1186/1758-2946-3-33",
"doi-asserted-by": "publisher",
"key": "CIT0040"
},
{
"DOI": "10.1063/1.328693",
"doi-asserted-by": "publisher",
"key": "CIT0041"
},
{
"author": "Pollard H. B.",
"journal-title": "bioRxiv",
"key": "CIT0042",
"year": "2020"
},
{
"author": "Rismanbaf A.",
"issue": "1",
"journal-title": "Archives of Academic Emergency Medicine",
"key": "CIT0043",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.2533/chimia.2007.312",
"doi-asserted-by": "publisher",
"key": "CIT0044"
},
{
"DOI": "10.1021/ci500588j",
"doi-asserted-by": "publisher",
"key": "CIT0045"
},
{
"DOI": "10.1016/0166-3542(96)00942-4",
"doi-asserted-by": "publisher",
"key": "CIT0046"
},
{
"DOI": "10.1107/S0907444904011679",
"doi-asserted-by": "publisher",
"key": "CIT0047"
},
{
"DOI": "10.20944/preprints202002.0418.v2",
"doi-asserted-by": "crossref",
"key": "CIT0048",
"unstructured": "Sekhar, T. (2020). Virtual screening based prediction of potential drugs for COVID-19. Preprints.org. https://doi.org/10.20944/preprints202002.0418.v2"
},
{
"DOI": "10.1038/s41586-020-2179-y",
"doi-asserted-by": "publisher",
"key": "CIT0049"
},
{
"DOI": "10.3390/pathogens9020148",
"doi-asserted-by": "publisher",
"key": "CIT0050"
},
{
"DOI": "10.3748/wjg.v22.i29.6638",
"doi-asserted-by": "publisher",
"key": "CIT0051"
},
{
"DOI": "10.1080/07391102.2018.1468281",
"doi-asserted-by": "publisher",
"key": "CIT0052"
},
{
"DOI": "10.1080/10799893.2017.1414844",
"doi-asserted-by": "publisher",
"key": "CIT0053"
},
{
"DOI": "10.1159/000149390",
"doi-asserted-by": "publisher",
"key": "CIT0054"
},
{
"DOI": "10.1371/journal.ppat.1007236",
"doi-asserted-by": "publisher",
"key": "CIT0055"
},
{
"DOI": "10.1136/gutjnl-2014-308971",
"doi-asserted-by": "publisher",
"key": "CIT0056"
},
{
"DOI": "10.1016/j.antiviral.2013.06.002",
"doi-asserted-by": "publisher",
"key": "CIT0057"
},
{
"DOI": "10.15406/japlr.2017.05.00156",
"doi-asserted-by": "publisher",
"key": "CIT0058"
},
{
"author": "Tinku Gupta M. M.",
"first-page": "58",
"issue": "1",
"journal-title": "Asian Journal of Pharmaceutical Education and Research",
"key": "CIT0059",
"volume": "7",
"year": "2018"
},
{
"DOI": "10.1016/0165-2478(94)00183-R",
"doi-asserted-by": "publisher",
"key": "CIT0060"
},
{
"DOI": "10.1128/AAC.41.3.551",
"doi-asserted-by": "publisher",
"key": "CIT0061"
},
{
"DOI": "10.1016/S1388-1981(99)00149-3",
"doi-asserted-by": "publisher",
"key": "CIT0062"
},
{
"key": "CIT0063",
"unstructured": "Visualizer, B. D. S. (2017). Version 4.5 (software). http://mayavi.sourceforge.net"
},
{
"DOI": "10.1042/BJ20120150",
"doi-asserted-by": "publisher",
"key": "CIT0064"
},
{
"DOI": "10.1126/science.abb2507",
"doi-asserted-by": "publisher",
"key": "CIT0065"
},
{
"DOI": "10.1038/s41586-020-2008-3",
"doi-asserted-by": "publisher",
"key": "CIT0066"
},
{
"DOI": "10.1007/s11427-020-1637-5",
"doi-asserted-by": "publisher",
"key": "CIT0067"
},
{
"DOI": "10.3390/molecules18066230",
"doi-asserted-by": "publisher",
"key": "CIT0068"
},
{
"DOI": "10.1038/s41421-020-0153-3",
"doi-asserted-by": "publisher",
"key": "CIT0069"
},
{
"DOI": "10.1056/NEJMoa2001017",
"doi-asserted-by": "publisher",
"key": "CIT0070"
}
],
"reference-count": 70,
"references-count": 70,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.tandfonline.com/doi/full/10.1080/07391102.2020.1824816"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Molecular Biology",
"General Medicine",
"Structural Biology"
],
"subtitle": [],
"title": "Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1080/tandf_crossmark_01",
"volume": "40"
}