Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach
Elvio Gayozo, Laura Rojas, Julio Barrios
Biotecnia, doi:10.18633/biotecnia.v27.2485
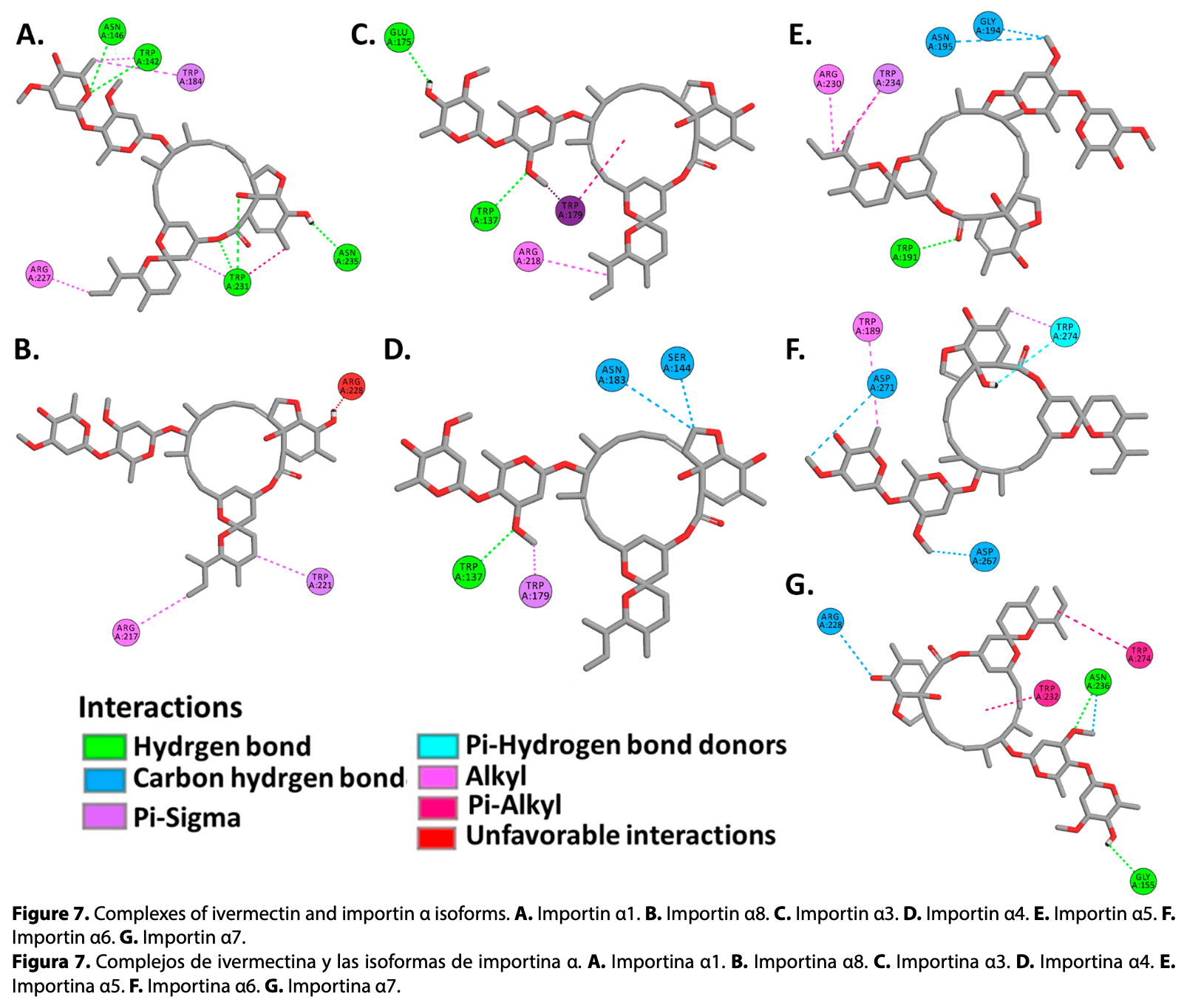
Ivermectin has been shown in vitro that reduces SARS-CoV-2 replication in infected cells through interactions with importins α, however, the exact mechanism of action is still unknown. The objective of this study was to analyze binding affinities of ivermectin, SARS-CoV-2 nucleocapsid (N) and ORF6 proteins, to isoforms of human importins α using molecular docking methods. Crystallized structures of importins α from Protein Data Bank (PDB) and AlphaFold Protein Structure Database were used, viral proteins were modeled using AlphaFold 2. Molecular docking simulations were performed between human importin α isoforms, ivermectin, N and ORF6 proteins, employing Broyden-Fletcher-Goldfarb-Shanno, FTDock and pyDockRST algorithms. Obtained data evidenced that viral proteins of SARS-CoV-2 and ivermectin showed favorable binding affinities to ARM2-ARM4 domains (major binding site), sharing binding affinities to the same active residues. These results suggest that ivermectin shares the same active site on the α-importins as the SARS-CoV-2 N and ORF6 proteins, demonstrating a potential molecular target for research in the development of new antiviral drugs against COVID-19.
Resulting complexes between importins α isoforms and N protein (NLS pat4 and pat7) showed free binding energy values (∆G) ranging -10.0 to -6.3 kcal.mol -1 , where the NLS pat7 sequence demonstrated the most favorable energy value, specifically to importin α3 and importin α5 (Figure 3 .B, Supplementary Table S1 ). Active residues in complexes formed by N protein and isoforms of α1 subfamily were Phe138, Trp142, Asn146, Ser149, His177, Glu180, Trp184, Asn188, Asn228 in importin α1, and Arg95, Gln100, Glu107, Trp136, Ser143, Glu180, Asn182, Trp231, Glu256, Asp260, Glu266, Asp270, Trp273 in importin α8 , residues that interact with NLS pat4 sequences through hydrogen bonds, unconventional interactions between a polarized carbon atom and hydrogen atom, interactions between Pi orbitals and donors groups of hydrogen bonds. The formation of electrostatic attractions and hydrophobic interactions were also identified (Figure 3 .A,B, Supplementary Figure S2 , Supplementary Figure S3 , Supplementary Table S1 ). Residues Glu107, Trp142, Asn146, Ala176, Glu180, Trp184, Ala222, Tyr225, Trp231, Glu266, Asp270, Trp273 of importin α1, and residues Ser99, Glu101, Pro104, Trp136, Ser143, Arg217, Asp260, Trp263, Glu174, Trp178, Asn182, Trp221, Asn225 of importin α8, interact by hydrogen bonds, carbon hydrogen bonds, Pi interactions with hydrogen bonds donors, hydrophobic and electrostatic interactions with the NLS pat7 sequences (Figure 4 .A,B, Supplementary Figure S2 , Supplementary..
References
Addetia, Lieberman, Phung, Hsiang, Xie et al., SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98, mBio,
doi:10.1128/mBio.00065-21Azam, Taban, Eid, Iqbal, Alam et al., An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin α, Journal of Biomolecular Structure and Dynamics,
doi:10.1080/07391102.2020.184102Bello, Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets, Journal of Biomolecular Structure and Dynamics,
doi:10.1080/07391102.2021.1911857Ceraolo, Giorgi, Genomic variance of the 2019-nCoV coronavirus, Journal of Medical Virology,
doi:10.1002/jmv.25700Chelliah, Blundell, Fernández-Recio, Efficient restraints for protein-protein docking by comparison of observed amino acid substitution patterns with those predicted from local Environment, Journal of Molecular Biology,
doi:10.1016/j.jmb.2006.01.001Chen, Boon, Wang, Chan, Chan, Genomic and evolutionary comparison between SARS-CoV-2 and other human coronaviruses, Journal of Virological Methods,
doi:10.1016/j.jviromet.2020.114032Cheng, Blundell, Fernandez-Recio, pyDock: Electrostatics and desolvation for effective scoring of rigid-body protein-protein docking, Proteins: Structure, Function, and Bioinformatics,
doi:10.1002/prot.21419Choudhury, Das, Patra, Bhattacharya, Ghosh et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach, Future Virology,
doi:10.2217/fvl-2020-0342Crump, Ivermectin: enigmatic multifaceted 'wonder' drug continues to surprise and exceed expectations, The Journal of Antibiotics,
doi:10.1038/ja.2017David, Islam, Tankhilevich, Sternberg, The AlphaFold database of protein structures: A Biologist's Guide, Journal of Molecular Biology,
doi:10.1016/j.jmb.2021.167336Fagerlund, Melén, Kinnunen, Julkunen, Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and Importin α5, Journal of Biological Chemistry,
doi:10.1074/jbc.M202943200Fang, Jang, Watson, Wellappili, Tyler, Distinctive nuclear localization signals in the oomycete Phytophthora sojae, Front. Microbiol,
doi:10.3389/fmicb.2017.00010Fontes, Teh, Kobe, Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α11, Journal of Molecular Biology,
doi:10.1006/jmbi.2000.3642Frieman, Yount, Heise, Kopecky-Bromberg, Palese et al., Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane, Journal of Virology,
doi:10.1128/JVI.01012-07Fry, Saladi, Cunha, Clemons, Sequence-based features that are determinant for tailanchored membrane protein sorting in eukaryotes, Traffic,
doi:10.1111/tra.12809Gabb, Jackson, Sternberg, Modelling protein docking using shape complementarity, electrostatics and biochemical information, J. Thornton. Journal of Molecular Biology,
doi:10.1006/jmbi.1997.1203Gao, Gao, Liu, Nie, Sun et al., Identification and functional analysis of the SARS-COV-2 nucleocapsid protein, BMC Microbiology,
doi:10.1186/s12866-021-02107-3González-Paz, Hurtado-León, Lossada, Fernández-Materán, Vera-Villalobos et al., Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach, Biophysical Chemistry,
doi:10.1016/j.bpc.2021.106677Gordon, Jang, Bouhaddou, Xu, Obernier et al., None
Grosdidier, Pons, Solernou, Fernández-Recio, Prediction and scoring of docking poses with pyDock, Proteins: Structure, Function, and Bioinformatics,
doi:10.1002/prot.21796Gupta, Biswal, Panda, Ray, Rana, Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin, Journal of Biomolecular Structure and Dynamics,
doi:10.1080/07391102.2020.1839564Hanwell, Curtis, Lonie, Vandermeersch, Zurek et al., Avogadro: an advanced semantic chemical editor, visualization, and analysis platform, J. Cheminform,
doi:10.1186/1758-2946-4-17He, Dobie, Ballantine, Leeson, Li et al., Analysis of multimerization of the SARS coronavirus nucleocapsid protein, Biochemical and Biophysical Research Communications,
doi:10.1016/j.bbrc.2004.02.074Heo, Lee, Seok, GalaxyRefineComplex: Refinement of protein-protein complex model structures driven by interface repacking, Sci Rep,
doi:10.1038/srep32153Horton, Park, Obayashi, Fujita, Harada et al., WoLF PSORT: protein localization predictor, Nucleic Acids Res,
doi:10.1093/nar/gkm259Ibrahim, Abdelmalek, Elshahat, Elfiky, COVID-19 spike-host cell receptor GRP78 binding site prediction, Journal of Infection,
doi:10.1016/j.jinf.2020.02.026Iqbal, Romero-Castillo, Bilal, Parra-Saldivar, The emergence of novel-coronavirus and its replication cycle -An overview, Journal of Pure and Applied Microbiology,
doi:10.22207/JPAM.14.1.03Jiménez-García, Pons, Fernández-Recio, pyDockWEB: a web server for rigid-body protein-protein docking using electrostatics and desolvation scoring, Bioinformatics,
doi:10.1093/bioinformatics/btt262Kato, Ikliptikawati, Kobayashi, Kondo, Lim et al., Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex, Biochemical and Biophysical Research Communications,
doi:10.1016/j.bbrc.2020.11.115Kim, Thiessen, Bolton, Chen, Fu et al., PubChem substance and compound databases, Nucleic Acids Res,
doi:10.1093/nar/gkv951King, Tessier, Dodge, Weinberg, Mymryk, Inhibition of human adenovirus replication by the importin α/β1 nuclear import inhibitor ivermectin, Journal of Virology,
doi:10.1128/JVI.00710-20Kopecky-Bromberg, Martínez-Sobrido, Frieman, Baric, Palese, Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists, Journal of Virology,
doi:10.1128/JVI.01782-06Kumar, Stecher, Li, Knyaz, Tamura, MEGA X: Molecular evolutionary genetics analysis across computing platforms, Mol Biol Evol,
doi:10.1093/molbev/msy096Laskowski, Macarthur, Moss, Thornton, PROCHECK: a program to check the stereochemical quality of protein structures, J. Appl. Cryst,
doi:10.1107/S0021889892009944Li, Cowley, Uludag, Gur, Mcwilliam et al., The EMBL-EBI bioinformatics web and programmatic tools framework, Nucleic Acids Res,
doi:10.1093/nar/gkv279Li, Liao, Wang, Tan, Luo et al., The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway, Virus Research,
doi:10.1016/j.virusres.2020.198074Lu, Zhao, Li, Niu, Yang et al., Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, The Lancet,
doi:10.1016/S0140-6736(20)30251-8Mastrangelo, Pezzullo, De Burghgraeve, Kaptein, Pastorino et al., Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug, J. Antimicrob Chemother,
doi:10.1093/jac/dks147Miyamoto, Itoh, Suzuki, Tanaka, Sakai et al., SARS-CoV-2 ORF6 disrupts nucleocytoplasmic trafficking to advance viral replication, Commun Biol,
doi:10.1038/s42003-022-03427-4Pallara, Jiménez-García, Romero, Moal, Fernández-Recio, pyDock scoring for the new modeling challenges in docking: Protein-peptide, homomultimers, and domain-domain interactions, Proteins: Structure, Function, and Bioinformatics,
doi:10.1002/prot.25184Pettersen, Goddard, Huang, Couch, Greenblatt et al., UCSF Chimera-A visualization system for exploratory research and analysis, Journal of Computational Chemistry,
doi:10.1002/jcc.20084Pumroy, Cingolani, Diversification of importin-α isoforms in cellular trafficking and disease states, Biochemical Journal,
doi:10.1042/BJ20141186Pumroy, Ke, Hart, Zachariae, Cingolani, Molecular determinants for nuclear import of influenza A PB2 by importin α isoforms 3 and 7, Structure,
doi:10.1016/j.str.2014.11.015Qinfen, Jinming, Xiaojun, Huanying, Jicheng et al., The life cycle of SARS coronavirus in Vero E6 cells, Journal of Medical Virology,
doi:10.1002/jmv.20095Rizzo, Ivermectin, antiviral properties and COVID-19: a possible new mechanism of action, Naunyn-Schmiedeberg's Arch Pharmacol,
doi:10.1007/s00210-020-01902-5Robert, Gouet, Deciphering key features in protein structures with the new ENDscript server, Nucleic Acids Res,
doi:10.1093/nar/gku316Rowland, Chauhan, Fang, Pekosz, Kerrigan et al., Intracellular localization of the severe acute respiratory syndrome Ccoronavirus nucleocapsid protein: Absence of nucleolar accumulation during infection and after expression as a recombinant protein in Vero cells, Journal of Virology,
doi:10.1128/JVI.79.17.11507-11512.2005Sekimoto, Imamoto, Nakajima, Hirano, Yoneda, Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1, The EMBO Journal,
doi:10.1093/emboj/16.23.7067Shereen, Khan, Kazmi, Bashir, Siddique, COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses, Journal of Advanced Research,
doi:10.1016/j.jare.2020.03.005Surjit, Liu, Jameel, Chow, Lal, The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors, Biochem J,
doi:10.1042/BJ20040984Tarendeau, Boudet, Guilligay, Mas, Bougault et al., Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit, Nat Struct Mol Biol,
doi:10.1038/nsmb1212Timani, Liao, Ye, Linbai, Zeng et al., Nuclear/ nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus, Virus Research,
doi:10.1016/j.virusres.2005.05.007Trott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, Journal of Computational Chemistry,
doi:10.1002/jcc.21334Vangone, Schaarschmidt, Koukos, Geng, Citro et al., Large-scale prediction of binding affinity in protein-small ligand complexes: the PRODIGY-LIG web server, Bioinformatics,
doi:10.1093/bioinformatics/bty816Varadi, Anyango, Deshpande, Nair, Natassia et al., AlphaFold Protein Structure Database: massively expanding the structural coverage of proteinsequence space with high-accuracy models, Nucleic Acids Research,
doi:10.1093/nar/gkab1061Wagstaff, Sivakumaran, Heaton, Harrich, Jans, Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus, Biochemical Journal,
doi:10.1042/BJ20120150Williams, Headd, Moriarty, Prisant, Videau et al., MolProbity: More and better reference data for improved all-atom structure validation, Protein Science,
doi:10.1002/pro.3330Wulan, Heydet, Walker, Gahan, Ghildyal, Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses, Front. Microbiol,
doi:10.3389/fmicb.2015.00553Wurm, Chen, Hodgson, Britton, Brooks et al., Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division, Journal of Virology,
doi:10.1128/JVI.75.19.9345-9356.2001Xu, Edwards, Borek, Feagins, Mittal et al., Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1, Cell Host & Microbe,
doi:10.1016/j.chom.2014.07.008Yang, Atkinson, Wang, Lee, Bogoyevitch et al., The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer, Antiviral Research,
doi:10.1016/j.antiviral.2020.104760Ye, Wong, Li, Xie, A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNKdependent apoptosis, Biochimica et Biophysica Acta (BBA) -General Subjects,
doi:10.1016/j.bbagen.2008.07.009Zhao, Falcón, Zhou, Netland, Enjuanes et al., Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication, Journal of Virology,
doi:10.1128/JVI.02371-08Zhou, Yang, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature,
doi:10.1038/s41586-020-2012-7DOI record:
{
"DOI": "10.18633/biotecnia.v27.2485",
"ISSN": [
"1665-1456",
"1665-1456"
],
"URL": "http://dx.doi.org/10.18633/biotecnia.v27.2485",
"abstract": "<jats:p>Ivermectin has been shown in vitro that reduces SARS-CoV-2 replication in infected cells through interactions with importins α, however, the exact mechanism of action is still unknown. The objective of this study was to analyze binding affinities of ivermectin, SARS-CoV-2 nucleocapsid (N) and ORF6 proteins, to isoforms of human importins α using molecular docking methods. Crystallized structures of importins α from Protein Data Bank (PDB) and AlphaFold Protein Structure Database were used, viral proteins were modeled using AlphaFold 2. Molecular docking simulations were performed between human importin α isoforms, ivermectin, N and ORF6 proteins, employing Broyden-Fletcher-Goldfarb-Shanno, FTDock and pyDockRST algorithms. Data obtained evidenced that viral proteins of SARS-CoV-2 and ivermectin showed favorable binding affinities to ARM2-ARM4 domains (major binding site), sharing binding affinities to the same active residues. These results suggest that ivermectin shares the same active site on the α-importins as the SARS-CoV-2 N and ORF6 proteins, demonstrating a potential molecular target for research in the development of new antiviral drugs against COVID-19.</jats:p>",
"author": [
{
"ORCID": "https://orcid.org/0000-0001-9309-7056",
"affiliation": [],
"authenticated-orcid": false,
"family": "Gayozo",
"given": "Elvio",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0002-8341-7746",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rojas",
"given": "Laura",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4120-4141",
"affiliation": [],
"authenticated-orcid": false,
"family": "Barrios",
"given": "Julio",
"sequence": "additional"
}
],
"container-title": "Biotecnia",
"container-title-short": "BIOTECNIA",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
3,
18
]
],
"date-time": "2025-03-18T20:53:10Z",
"timestamp": 1742331190000
},
"deposited": {
"date-parts": [
[
2025,
3,
18
]
],
"date-time": "2025-03-18T20:53:30Z",
"timestamp": 1742331210000
},
"indexed": {
"date-parts": [
[
2025,
3,
19
]
],
"date-time": "2025-03-19T04:33:22Z",
"timestamp": 1742358802766,
"version": "3.40.1"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
3,
13
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc-sa/4.0",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
3,
13
]
],
"date-time": "2025-03-13T00:00:00Z",
"timestamp": 1741824000000
}
}
],
"link": [
{
"URL": "https://www.biotecnia.unison.mx/index.php/biotecnia/article/download/2485/1391",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.biotecnia.unison.mx/index.php/biotecnia/article/download/2485/1392",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.biotecnia.unison.mx/index.php/biotecnia/article/download/2485/1391",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "7961",
"original-title": [],
"page": "e2485",
"prefix": "10.18633",
"published": {
"date-parts": [
[
2025,
3,
13
]
]
},
"published-online": {
"date-parts": [
[
2025,
3,
13
]
]
},
"publisher": "Universidad de Sonora",
"reference": [
{
"DOI": "10.1128/mBio.00065-21",
"doi-asserted-by": "crossref",
"key": "75797",
"unstructured": "Addetia, A., Lieberman, N. A. P., Phung, Q., Hsiang, T.Y., Xie, H., Roychoudhury, P., Shrestha, L., Loprieno, M. A, Huang, M. L., Gale, M. J., Jerome, K. R. and Greninger, A. L. 2021. SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98. mBio, 12(2), e00065-21. https://doi.org/10.1128/mBio.00065-21"
},
{
"DOI": "10.1038/s41591-020-0820-9",
"doi-asserted-by": "crossref",
"key": "75798",
"unstructured": "Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C. and Garry, R. F. 2020. The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450-452. https://doi.org/10.1038/s41591-020-0820-9"
},
{
"DOI": "10.1080/07391102.2020.1841028",
"doi-asserted-by": "crossref",
"key": "75799",
"unstructured": "Azam, F., Taban, I. M., Eid, E. E. M., Iqbal, M., Alam, O., Khan, S., Mahmood, D., Anwar, M. J., Khaililullah, H. and Khan, M. U. 2020. An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin α. Journal of Biomolecular Structure and Dynamics, 40(6), 2851–2864. https://doi.org/10.1080/07391102.2020.1841028"
},
{
"DOI": "10.1007/978-3-319-77309-4_6",
"doi-asserted-by": "crossref",
"key": "75800",
"unstructured": "Baumhardt, J. and Chook, Y. M. 2018. Structures of Importins and Exportins. En W. Yang (Ed.), Nuclear-Cytoplasmic Transport (pp. 113-149). Cham: Springer International Publishing. https://doi.org/10.1007/978-3-319-77309-4_6"
},
{
"DOI": "10.1080/07391102.2021.1911857",
"doi-asserted-by": "crossref",
"key": "75801",
"unstructured": "Bello, M. 2022. Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets. Journal of Biomolecular Structure and Dynamics, 40(18), 8375-8383. https://doi.org/10.1080/07391102.2021.1911857"
},
{
"DOI": "10.1093/nar/28.1.235",
"doi-asserted-by": "crossref",
"key": "75802",
"unstructured": "Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. and Bourne, P. E. 2000. The Protein Data Bank. Nucleic Acids Research, 28(1), 235-242. https://doi.org/10.1093/nar/28.1.235"
},
{
"DOI": "10.1016/j.virusres.2010.02.011",
"doi-asserted-by": "crossref",
"key": "75803",
"unstructured": "Bian, X.L. and Wilson, V. G. 2010. Common importin alpha specificity for papillomavirus E2 proteins. Virus Research, 150(1), 135-137. https://doi.org/10.1016/j.virusres.2010.02.011"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"doi-asserted-by": "crossref",
"key": "75804",
"unstructured": "Caly, L., Druce, J. D., Catton, M. G., Jans, D. A. and Wagstaff, K. M. 2020. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, 178, 104787. https://doi.org/10.1016/j.antiviral.2020.104787"
},
{
"DOI": "10.1002/jmv.25700",
"doi-asserted-by": "crossref",
"key": "75805",
"unstructured": "Ceraolo, C. and Giorgi, F. M. 2020. Genomic variance of the 2019-nCoV coronavirus. Journal of Medical Virology, 92(5), 522-528. https://doi.org/10.1002/jmv.25700"
},
{
"DOI": "10.1016/j.antiviral.2013.12.009",
"doi-asserted-by": "crossref",
"key": "75806",
"unstructured": "Chang, C., Hou, M.H., Chang, C.F., Hsiao, C.D. and Huang, T. 2014. The SARS coronavirus nucleocapsid protein – Forms and functions. Antiviral Research, 103, 39-50. https://doi.org/10.1016/j.antiviral.2013.12.009"
},
{
"DOI": "10.1016/j.jmb.2006.01.001",
"doi-asserted-by": "crossref",
"key": "75807",
"unstructured": "Chelliah, V., Blundell, T. L. and Fernández-Recio, J. 2006. Efficient Restraints for Protein–Protein Docking by Comparison of Observed Amino Acid Substitution Patterns with those Predicted from Local Environment. Journal of Molecular Biology, 357(5), 1669-1682. https://doi.org/10.1016/j.jmb.2006.01.001"
},
{
"DOI": "10.1002/prot.21419",
"doi-asserted-by": "crossref",
"key": "75808",
"unstructured": "Cheng, T. M.K., Blundell, T. L. and Fernandez-Recio, J. 2007. pyDock: Electrostatics and desolvation for effective scoring of rigid-body protein–protein docking. Proteins: Structure, Function, and Bioinformatics, 68(2), 503-515. https://doi.org/10.1002/prot.21419"
},
{
"DOI": "10.1016/S0959-440X(01)00264-0",
"doi-asserted-by": "crossref",
"key": "75809",
"unstructured": "Chook, Y. and Blobel, G. 2001. Karyopherins and nuclear import. Current Opinion in Structural Biology, 11(6), 703-715. https://doi.org/10.1016/S0959-440X(01)00264-0"
},
{
"DOI": "10.2217/fvl-2020-0342",
"doi-asserted-by": "crossref",
"key": "75810",
"unstructured": "Choudhury, A., Das, N. C., Patra, R., Bhattacharya, M., Ghosh, P., Patra, B. C. and Mukherjee, S. 2021. Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: An in silico approach. Future Virology, 16(4), 277-291. https://doi.org/10.2217/fvl-2020-0342"
},
{
"DOI": "10.1038/ja.2017.11",
"doi-asserted-by": "crossref",
"key": "75811",
"unstructured": "Crump, A. 2017. Ivermectin: Enigmatic multifaceted ‘wonder’ drug continues to surprise and exceed expectations. The Journal of Antibiotics, 70(5), 495-505. https://doi.org/10.1038/ja.2017.11"
},
{
"DOI": "10.1007/978-1-4939-2269-7_19",
"doi-asserted-by": "crossref",
"key": "75812",
"unstructured": "Dallakyan, S. and Olson, A. J. 2015. Small-Molecule Library Screening by Docking with PyRx. En J. E. Hempel, C. H. Williams, and C. C. Hong (Eds.), Chemical Biology: Methods and Protocols (pp. 243-250). New York, NY: Springer. https://doi.org/10.1007/978-1-4939-2269-7_19"
},
{
"DOI": "10.1016/j.jmb.2021.167336",
"doi-asserted-by": "crossref",
"key": "75813",
"unstructured": "David, A., Islam, S., Tankhilevich, E. and Sternberg, M. J. E. 2022. The AlphaFold Database of Protein Structures: A Biologist’s Guide. Journal of Molecular Biology, 434(2), 167336. https://doi.org/10.1016/j.jmb.2021.167336"
},
{
"DOI": "10.1074/jbc.M202943200",
"doi-asserted-by": "crossref",
"key": "75814",
"unstructured": "Fagerlund, R., Melén, K., Kinnunen, L. and Julkunen, I. 2002. Arginine/Lysine-rich Nuclear Localization Signals Mediate Interactions between Dimeric STATs and Importin α5 *. Journal of Biological Chemistry, 277(33), 30072-30078. https://doi.org/10.1074/jbc.M202943200"
},
{
"DOI": "10.1006/jmbi.2000.3642",
"doi-asserted-by": "crossref",
"key": "75815",
"unstructured": "Fontes, M. R. M., Teh, T. and Kobe, B. 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α11Edited by K. Nagai. Journal of Molecular Biology, 297(5), 1183-1194. https://doi.org/10.1006/jmbi.2000.3642"
},
{
"DOI": "10.1128/JVI.01012-07",
"doi-asserted-by": "crossref",
"key": "75816",
"unstructured": "Frieman, M., Yount, B., Heise, M., Kopecky-Bromberg, S. A., Palese, P. and Baric, R. S. 2007. Severe Acute Respiratory Syndrome Coronavirus ORF6 Antagonizes STAT1 Function by Sequestering Nuclear Import Factors on the Rough Endoplasmic Reticulum/Golgi Membrane. Journal of Virology, 81(18), 9812-9824. https://doi.org/10.1128/JVI.01012-07"
},
{
"DOI": "10.1006/jmbi.1997.1203",
"doi-asserted-by": "crossref",
"key": "75817",
"unstructured": "Gabb, H. A., Jackson, R. M. and Sternberg, M. J. E. 1997. Modelling protein docking using shape complementarity, electrostatics and biochemical information11Edited by J. Thornton. Journal of Molecular Biology, 272(1), 106-120. https://doi.org/10.1006/jmbi.1997.1203"
},
{
"DOI": "10.1186/s12866-021-02107-3",
"doi-asserted-by": "crossref",
"key": "75818",
"unstructured": "Gao, T., Gao, Y., Liu, X., Nie, Z., Sun, H., Lin, K., Peng, H. and Wang, S. 2021. Identification and functional analysis of the SARS-COV-2 nucleocapsid protein. BMC Microbiology, 21(1), 58. https://doi.org/10.1186/s12866-021-02107-3"
},
{
"DOI": "10.56152/ffs.v12i2.2219",
"doi-asserted-by": "crossref",
"key": "75819",
"unstructured": "Gayozo, E., Rojas, L. and López, M. 2020. Análisis computacional de la interacción in silico de la Agatisflavona con la región de trimerización de la proteína espícula (S) del SARS-CoV-2. Steviana, 12(2), 70-80. https://doi.org/10.56152/StevianaFacenV12N2A4_2020"
},
{
"DOI": "10.15446/rev.colomb.biote.v23n2.94466",
"doi-asserted-by": "crossref",
"key": "75820",
"unstructured": "Gayozo, E. and Rojas, L. 2021. Interacción in silico de las moléculas Agathisflavona, Amentoflavona y Punicalina con la Importina α1 humana. Revista Colombiana de Biotecnología, 23(2), 15-24. https://doi.org/10.15446/rev.colomb.biote.v23n2.94466"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"doi-asserted-by": "crossref",
"key": "75821",
"unstructured": "Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., O’Meara, M. J., Rezelj, V. V., Guo, J. Z., Swaney, D. L., Tummino, T. A., Hüttenhain, R., Kaake, R. M., Richards, A. L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., Kim, M., Haas, P., Polacco, B. J., Braberg, H., Fabius, J. M., Eckhardt, M., Soucheray, M., Bennett, M. J., Cakir, M., McGregor, M. J., Li, Q., Meyer, B., Roesch, F., Vallet, T., Mac Kain, A., Miorin, L., Moreno, E., Chi Naing, Z. Z., Zhou, Y., Peng, S., Shi, Y., Zhang, Z., Shen, W., Kirby, I. T., Melnyk, J. E., Chorba, J. S., Lou, K., Dai, S. A., Barrio-Hernandez, I., Memon, D., Hernandez-Armenta, C., Lyu, J., Mathy, C. P., Perica, T., Bharath Pilla, K., Ganesan, S. J., Saltzberg, D. J., Rakesh, R., Liu, X., Rosenthal, S. B., Calviello, L., Venkataramanan, S., Liboy-Lugo, J., Lin, Y., Huang, X. P., Liu, Y., Wankowicz, S. A., Bohn, M., Safari, M., Ugur, F. S., Koh, C., Savar, N. S., Dinh Tran, Q., Shengjuler,D., Fletcher, S. J., O’Neal, M. C., Cai, Y., Chang, J. C., Broadhurst, D. J., Klippsten, S., Sharp, P. P., Wenzell, N. A., Kuzuoglu-Ozturk, D., Wang, H., Trenker, R., Young, J. M., Cavero, D. A., Hiatt, J., Roth, T. L., Rathore, U., Subramanian, A., Noack, J., Hubert, M., Stroud, R. M., Frankel, A. D., Rosenberg, O. S., Verba, K. A., Agard, D. A., Ott, M., Emerman, M., Jura, N., von Zastrow, M., Verdin, E., Ashworth, A., Schwartz, O., d’Enfert, C., Mukherjee, S., Jacobson, M., Malik, H. M., Fujimori, D. G., Ideker, T., Craik, C. S., Floor, S. N., Fraser, J. S., Gross, J. D., Sali, A., Roth, B. L., Ruggero, D., Taunton, J., Kortemme, T., Beltrao, P., Vignuzzi, M., García-Sastre, A., Shokat, K. M., Shoichet, B. K. and Krogan, N. J. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816), 459-468. https://doi.org/10.1038/s41586-020-2286-9"
},
{
"DOI": "10.1002/prot.21796",
"doi-asserted-by": "crossref",
"key": "75822",
"unstructured": "Grosdidier, S., Pons, C., Solernou, A. and Fernández-Recio, J. 2007. Prediction and scoring of docking poses with pyDock. Proteins: Structure, Function, and Bioinformatics, 69(4), 852-858. https://doi.org/10.1002/prot.21796"
},
{
"DOI": "10.1016/j.bbrc.2004.02.074",
"doi-asserted-by": "crossref",
"key": "75823",
"unstructured": "He, R., Dobie, F., Ballantine, M., Leeson, A., Li, Y., Bastien, N., Cutts, T., Andonov, A., Cao, J., Booth, T. F., Plummer, F. A., Tyler, S., Baker, L. and Li, X. 2004. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochemical and Biophysical Research Communications, 316(2), 476-483. https://doi.org/10.1016/j.bbrc.2004.02.074"
},
{
"DOI": "10.1038/srep32153",
"doi-asserted-by": "crossref",
"key": "75824",
"unstructured": "Heo, L., Lee, H. and Seok, C. 2016. GalaxyRefineComplex: Refinement of protein-protein complex model structures driven by interface repacking. Scientific Reports, 6(1), 32153. https://doi.org/10.1038/srep32153"
},
{
"DOI": "10.1093/nar/gkm259",
"doi-asserted-by": "crossref",
"key": "75825",
"unstructured": "Horton, P., Park, K.J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J. and Nakai, K. 2007. WoLF PSORT: Protein localization predictor. Nucleic Acids Research, 35(suppl_2), W585-W587. https://doi.org/10.1093/nar/gkm259"
},
{
"DOI": "10.1016/j.virusres.2010.08.017",
"doi-asserted-by": "crossref",
"key": "75826",
"unstructured": "Hussain, S. and Gallagher, T. 2010. SARS-coronavirus protein 6 conformations required to impede protein import into the nucleus. Virus Research, 153(2), 299-304. https://doi.org/10.1016/j.virusres.2010.08.017"
},
{
"DOI": "10.22207/JPAM.14.1.03",
"doi-asserted-by": "crossref",
"key": "75827",
"unstructured": "Iqbal, H. M. N., Romero-Castillo, K. D., Bilal, M. and Parra-Saldivar, R. 2020. The emergence of novel-coronavirus and its replication cycle—An overview. Journal of Pure and Applied Microbiology, 14(1). https://doi.org/10.22207/JPAM.14.1.03"
},
{
"DOI": "10.1093/bioinformatics/btt262",
"doi-asserted-by": "crossref",
"key": "75828",
"unstructured": "Jiménez-García, B., Pons, C. and Fernández-Recio, J. 2013. pyDockWEB: A web server for rigid-body protein–protein docking using electrostatics and desolvation scoring. Bioinformatics, 29(13), 1698-1699. https://doi.org/10.1093/bioinformatics/btt262"
},
{
"DOI": "10.1038/s41586-021-03819-2",
"doi-asserted-by": "crossref",
"key": "75829",
"unstructured": "Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A., Potapenko, A., Bridgland, A., Meyer, C., Kohl, S. A., Ballard, A. J., Cowie, A., Romera-Paredes, B., Nikolov, S., Jain, R., Adler, J., Back, T., Petersen, S., Reiman, D., Clancy, E., Zielinski, M., Steinegger, M., Pacholska, M., Berghammer, T., Bodenstein, S., Silver, D., Vinyals, O., Senior, A. W., Kavukcuoglu, K., Kohli, P. and Hassabis, D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873), 583-589. https://doi.org/10.1038/s41586-021-03819-2"
},
{
"key": "75830",
"unstructured": "Kannan, S., Shaik Syed Ali, P., Sheeza, A. and Hemalatha, K. 2020. COVID-19 (Novel Coronavirus 2019)—Recent trends. European Review for Medical and Pharmacological Sciences, 24(4), 2006-2011. https://doi.org/10.26355/eurrev_202002_20378"
},
{
"DOI": "10.1016/j.bbrc.2020.11.115",
"doi-asserted-by": "crossref",
"key": "75831",
"unstructured": "Kato, K., Ikliptikawati, D. K., Kobayashi, A., Kondo, H., Lim, K., Hazawa, M. and Wong, R. W. 2021. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochemical and Biophysical Research Communications, 536, 59-66. https://doi.org/10.1016/j.bbrc.2020.11.115"
},
{
"DOI": "10.1093/nar/gkv951",
"doi-asserted-by": "crossref",
"key": "75832",
"unstructured": "Kim, S., Thiessen, P. A., Bolton, E. E., Chen, J., Fu, G., Gindulyte, A., Han, L., He, J., He, S., Shoemaker, B. A., Wang, J., Yu, B., Zhang, J. and Bryant, S. H. 2016. PubChem Substance and Compound databases. Nucleic Acids Research, 44(D1), D1202-D1213. https://doi.org/10.1093/nar/gkv951"
},
{
"DOI": "10.1128/JVI.00710-20",
"doi-asserted-by": "crossref",
"key": "75833",
"unstructured": "King, C. R., Tessier, T. M., Dodge, M. J., Weinberg, J. B. and Mymryk, J. S. 2020. Inhibition of Human Adenovirus Replication by the Importin α/β1 Nuclear Import Inhibitor Ivermectin. Journal of Virology, 94(18), e00710-20. https://doi.org/10.1128/JVI.00710-20"
},
{
"DOI": "10.1128/JVI.01782-06",
"doi-asserted-by": "crossref",
"key": "75834",
"unstructured": "Kopecky-Bromberg, S. A., Martínez-Sobrido, L., Frieman, M., Baric, R. A. and Palese, P. 2007. Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. Journal of Virology, 81(2), 548-557. https://doi.org/10.1128/JVI.01782-06"
},
{
"DOI": "10.1093/molbev/msy096",
"doi-asserted-by": "crossref",
"key": "75835",
"unstructured": "Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35(6), 1547-1549. https://doi.org/10.1093/molbev/msy096"
},
{
"DOI": "10.1016/0022-2836(82)90515-0",
"doi-asserted-by": "crossref",
"key": "75836",
"unstructured": "Kyte, J. and Doolittle, R. F. 1982. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology, 157(1), 105-132. https://doi.org/10.1016/0022-2836(82)90515-0"
},
{
"DOI": "10.1107/S0021889892009944",
"doi-asserted-by": "crossref",
"key": "75837",
"unstructured": "Laskowski, R. A., MacArthur, M. W., Moss, D. S. and Thornton, J. M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26(2), 283-291. https://doi.org/10.1107/S0021889892009944"
},
{
"DOI": "10.1016/j.virusres.2020.198074",
"doi-asserted-by": "crossref",
"key": "75838",
"unstructured": "Li, J.Y., Liao, C.H., Wang, Q., Tan, Y.J., Luo, R., Qiu, Y. and Ge, X.Y. 2020. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Research, 286, 198074. https://doi.org/10.1016/j.virusres.2020.198074"
},
{
"DOI": "10.1093/nar/gkv279",
"doi-asserted-by": "crossref",
"key": "75839",
"unstructured": "Li, W., Cowley, A., Uludag, M., Gur, T., McWilliam, H., Squizzato, S., Park, Y. M., Buso, N. and Lopez, R. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Research, 43(W1), W580-W584. https://doi.org/10.1093/nar/gkv279"
},
{
"DOI": "10.1016/j.antiviral.2014.06.013",
"doi-asserted-by": "crossref",
"key": "75840",
"unstructured": "Liu, D. X., Fung, T. S., Chong, K. K.L., Shukla, A. and Hilgenfeld, R. 2014. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Research, 109, 97-109. https://doi.org/10.1016/j.antiviral.2014.06.013"
},
{
"DOI": "10.1016/S0140-6736(20)30251-8",
"doi-asserted-by": "crossref",
"key": "75841",
"unstructured": "Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., Chen, J., Meng, Y., Wang, J., Lin, Y., Yuan, J., Xie, Z., Ma, J., Liu, W. L., Wang, D., Xu, W., Holmes, E. C., Gao, G. F., Wu, G., Chen, W., Shi, W. and Tan, W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565-574. https://doi.org/10.1016/S0140-6736(20)30251-8"
},
{
"DOI": "10.1093/jac/dks147",
"doi-asserted-by": "crossref",
"key": "75842",
"unstructured": "Mastrangelo, E., Pezzullo, M., De Burghgraeve, T., Kaptein, S., Pastorino, B., Dallmeier, K., de Lamballerie, X., Neyts, J., Hanson, A. M., Frick, D. N., Bolognesi, M. and Milani, M. 2012. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: New prospects for an old drug. Journal of Antimicrobial Chemotherapy, 67(8), 1884-1894. https://doi.org/10.1093/jac/dks147"
},
{
"DOI": "10.1038/s41592-022-01488-1",
"doi-asserted-by": "crossref",
"key": "75843",
"unstructured": "Mirdita, M., Schütze, K., Moriwaki, Y., Heo, L., Ovchinnikov, S. and Steinegger, M. 2022. ColabFold: Making protein folding accessible to all. Nature Methods, 19(6), 679-682. https://doi.org/10.1038/s41592-022-01488-1"
},
{
"DOI": "10.1038/s42003-022-03427-4",
"doi-asserted-by": "crossref",
"key": "75844",
"unstructured": "Miyamoto, Y., Itoh, Y., Suzuki, T., Tanaka, T., Sakai, Y., Koido, M., Hata, C., Wang, C. X., Otani, M., Moriishi, K., Tachibana, T., Kamatani, Y., Yoneda, Y., Okamoto, T. and Oka, M. 2022. SARS-CoV-2 ORF6 disrupts nucleocytoplasmic trafficking to advance viral replication. Communications Biology, 5(1), 1-15. https://doi.org/10.1038/s42003-022-03427-4"
},
{
"DOI": "10.1016/j.virusres.2007.10.009",
"doi-asserted-by": "crossref",
"key": "75845",
"unstructured": "Narayanan, K., Huang, C. and Makino, S. 2008. SARS coronavirus accessory proteins. Virus Research, 133(1), 113-121. https://doi.org/10.1016/j.virusres.2007.10.009"
},
{
"DOI": "10.1016/j.pt.2014.07.005",
"doi-asserted-by": "crossref",
"key": "75846",
"unstructured": "Ōmura, S. and Crump, A. 2014. Ivermectin: Panacea for resource-poor communities? Trends in Parasitology, 30(9), 445-455. https://doi.org/10.1016/j.pt.2014.07.005"
},
{
"DOI": "10.1002/prot.25184",
"doi-asserted-by": "crossref",
"key": "75847",
"unstructured": "Pallara, C., Jiménez-García, B., Romero, M., Moal, I. H. and Fernández-Recio, J. 2017. pyDock scoring for the new modeling challenges in docking: Protein–peptide, homo-multimers, and domain–domain interactions. Proteins: Structure, Function, and Bioinformatics, 85(3), 487-496. https://doi.org/10.1002/prot.25184"
},
{
"DOI": "10.1002/jcc.20084",
"doi-asserted-by": "crossref",
"key": "75848",
"unstructured": "Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. and Ferrin, T. E. 2004. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605-1612. https://doi.org/10.1002/jcc.20084"
},
{
"DOI": "10.1016/j.str.2014.11.015",
"doi-asserted-by": "crossref",
"key": "75849",
"unstructured": "Pumroy, R. A., Ke, S., Hart, D. J., Zachariae, U. and Cingolani, G. 2015. Molecular Determinants for Nuclear Import of Influenza A PB2 by Importin α Isoforms 3 and 7. Structure, 23(2), 374-384. https://doi.org/10.1016/j.str.2014.11.015"
},
{
"DOI": "10.1042/BJ20141186",
"doi-asserted-by": "crossref",
"key": "75850",
"unstructured": "Pumroy, R. and Cingolani, G. 2015. Diversification of importin-α isoforms in cellular trafficking and disease states. The Biochemical journal. https://doi.org/10.1042/BJ20141186"
},
{
"DOI": "10.1002/jmv.20095",
"doi-asserted-by": "crossref",
"key": "75851",
"unstructured": "Qinfen, Z., Jinming, C., Xiaojun, H., Huanying, Z., Jicheng, H., Ling, F., Kunpeng, L. and Jingqiang, Z. 2004. The life cycle of SARS coronavirus in Vero E6 cells. Journal of Medical Virology, 73(3), 332-337. https://doi.org/10.1002/jmv.20095"
},
{
"DOI": "10.1007/s00210-020-01902-5",
"doi-asserted-by": "crossref",
"key": "75852",
"unstructured": "Rizzo, E. 2020. Ivermectin, antiviral properties and COVID-19: A possible new mechanism of action. Naunyn-Schmiedeberg’s Archives of Pharmacology, 393(7), 1153-1156. https://doi.org/10.1007/s00210-020-01902-5"
},
{
"DOI": "10.1093/nar/gku316",
"doi-asserted-by": "crossref",
"key": "75853",
"unstructured": "Robert, X. and Gouet, P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42(W1), W320-W324. https://doi.org/10.1093/nar/gku316"
},
{
"DOI": "10.1128/JVI.79.17.11507-11512.2005",
"doi-asserted-by": "crossref",
"key": "75854",
"unstructured": "Rowland, R. R. R., Chauhan, V., Fang, Y., Pekosz, A., Kerrigan, M. and Burton, M. D. 2005. Intracellular Localization of the Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein: Absence of Nucleolar Accumulation during Infection and after Expression as a Recombinant Protein in Vero Cells. Journal of Virology, 79(17), 11507-11512. https://doi.org/10.1128/JVI.79.17.11507-11512.2005"
},
{
"DOI": "10.1007/s11224-021-01776-0",
"doi-asserted-by": "crossref",
"key": "75855",
"unstructured": "Saha, J. K. and Raihan, Md. J. 2021. The binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2. Structural Chemistry, 32(5), 1985-1992. https://doi.org/10.1007/s11224-021-01776-0"
},
{
"DOI": "10.1093/emboj/16.23.7067",
"doi-asserted-by": "crossref",
"key": "75856",
"unstructured": "Sekimoto, T., Imamoto, N., Nakajima, K., Hirano, T. and Yoneda, Y. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. The EMBO Journal, 16(23), 7067-7077. https://doi.org/10.1093/emboj/16.23.7067"
},
{
"DOI": "10.1080/07391102.2020.1839564",
"doi-asserted-by": "crossref",
"key": "75857",
"unstructured": "Sen Gupta, P. S., Biswal, S., Panda, S. K., Ray, A. K. and Rana, M. K. 2022. Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin. Journal of Biomolecular Structure and Dynamics, 40(5), 2217-2226. https://doi.org/10.1080/07391102.2020.1839564"
},
{
"DOI": "10.1016/j.jare.2020.03.005",
"doi-asserted-by": "crossref",
"key": "75858",
"unstructured": "Shereen, M. A., Khan, S., Kazmi, A., Bashir, N. and Siddique, R. 2020. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91-98. https://doi.org/10.1016/j.jare.2020.03.005"
},
{
"DOI": "10.1007/978-1-62703-646-7_6",
"doi-asserted-by": "crossref",
"key": "75859",
"unstructured": "Sievers, F. and Higgins, D. G. 2014. Clustal Omega, Accurate Alignment of Very Large Numbers of Sequences. En D. J. Russell (Ed.), Multiple Sequence Alignment Methods (pp. 105-116). Totowa, NJ: Humana Press. https://doi.org/10.1007/978-1-62703-646-7_6"
},
{
"DOI": "10.1042/BJ20040984",
"doi-asserted-by": "crossref",
"key": "75860",
"unstructured": "Surjit, M., Liu, B., Jameel, S., Chow, V. T. K. and Lal, S. K. 2004. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochemical Journal, 383(1), 13-18. https://doi.org/10.1042/BJ20040984"
},
{
"DOI": "10.1038/nsmb1212",
"doi-asserted-by": "crossref",
"key": "75861",
"unstructured": "Tarendeau, F., Boudet, J., Guilligay, D., Mas, P. J., Bougault, C. M., Boulo, S., Baudin, F., Ruigrok, R. W. H., Daigle, N., Ellenberg, J., Cusack, S., Simorre, J. P. and Hart, D. J. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nature Structural & Molecular Biology, 14(3), 229-233. https://doi.org/10.1038/nsmb1212"
},
{
"DOI": "10.1016/j.virusres.2005.05.007",
"doi-asserted-by": "crossref",
"key": "75862",
"unstructured": "Timani, K. A., Liao, Q., Ye, L., Zeng, Y., Liu, J., Zheng, Y., Ye, L., Yang, X., Lingbao, K., Gao, J. and Zhu, Y. 2005. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Research, 114(1), 23-34. https://doi.org/10.1016/j.virusres.2005.05.007"
},
{
"DOI": "10.1002/jcc.21334",
"doi-asserted-by": "crossref",
"key": "75863",
"unstructured": "Trott, O. and Olson, A. J. 2010. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455-461. https://doi.org/10.1002/jcc.21334"
},
{
"DOI": "10.1093/bioinformatics/bty816",
"doi-asserted-by": "crossref",
"key": "75864",
"unstructured": "Vangone, A., Schaarschmidt, J., Koukos, P., Geng, C., Citro, N., Trellet, M. E., Xue, L. C. and Bonvin, A. M. J. J. 2019. Large-scale prediction of binding affinity in protein–small ligand complexes: The PRODIGY-LIG web server. Bioinformatics, 35(9), 1585-1587. https://doi.org/10.1093/bioinformatics/bty816"
},
{
"DOI": "10.1093/nar/gkab1061",
"doi-asserted-by": "crossref",
"key": "75865",
"unstructured": "Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., Yuan, D., Stroe, O., Wood, G., Laydon, A., Žídek, A., Green, T., Tunyasuvunakool, K., Petersen, S., Jumper, J., Clancy, E., Green, R., Vora, A., Lutfi, M., Figurnov, M., Cowie, A., Hobbs, N., Kohli, P., Kleywegt, G., Birney, E., Hassabis, D. and Velankar, S. 2022. AlphaFold Protein Structure Database: Massively expanding the structural coverag2e of protein-sequence space with high-accuracy models. Nucleic Acids Research, 50(D1), D439-D444. https://doi.org/10.1093/nar/gkab1061"
},
{
"DOI": "10.1042/BJ20120150",
"doi-asserted-by": "crossref",
"key": "75866",
"unstructured": "Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D. and Jans, D. A. 2012. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochemical Journal, 443(3), 851-856. https://doi.org/10.1042/BJ20120150"
},
{
"DOI": "10.1002/pro.3330",
"doi-asserted-by": "crossref",
"key": "75867",
"unstructured": "Williams, C. J., Headd, J. J., Moriarty, N. W., Prisant, M. G., Videau, L. L., Deis, L. N., Verma, V., Keedy, D. A., Hintze, B. J., Chen, V. B., Jain, S., Lewis, S. M., Arendall III, W. B., Snoeyink, J., Adams, P. D., Lovell, S. C., Richardson, J. S. and Richardson, D. C. 2018. MolProbity: More and better reference data for improved all-atom structure validation. Protein Science, 27(1), 293-315. https://doi.org/10.1002/pro.3330"
},
{
"DOI": "10.1038/s41586-020-2008-3",
"doi-asserted-by": "crossref",
"key": "75868",
"unstructured": "Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., Hu, Y., Tao, Z. W., Tian, J. H., Pei, Y. Y., Yuan, M. L., Zhang, Y. L., Dai, F. H., Liu, Y., Wang, Q. M., Zheng, J. J., Xu, L., Holmes, E. C. and Zhang, Y. Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265-269. https://doi.org/10.1038/s41586-020-2008-3"
},
{
"DOI": "10.3389/fmicb.2015.00553",
"doi-asserted-by": "crossref",
"key": "75869",
"unstructured": "Wulan, W. N., Heydet, D., Walker, E. J., Gahan, M. E. and Ghildyal, R. 2015. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Frontiers in Microbiology, 6. https://doi.org/10.3389/fmicb.2015.00553"
},
{
"DOI": "10.1128/JVI.75.19.9345-9356.2001",
"doi-asserted-by": "crossref",
"key": "75870",
"unstructured": "Wurm, T., Chen, H., Hodgson, T., Britton, P., Brooks, G. and Hiscox, J. A. 2001. Localization to the Nucleolus Is a Common Feature of Coronavirus Nucleoproteins, and the Protein May Disrupt Host Cell Division. Journal of Virology, 75(19), 9345-9356. https://doi.org/10.1128/JVI.75.19.9345-9356.2001"
},
{
"DOI": "10.1016/j.chom.2014.07.008",
"doi-asserted-by": "crossref",
"key": "75871",
"unstructured": "Xu, W., Edwards, M. R., Borek, D. M., Feagins, A. R., Mittal, A., Alinger, J. B., Berry, K. N., Yen, B., Hamilton, J., Brett, T. J., Pappu, R. V., Leung, D. W., Basler, C. F. and Amarasinghe, G. K. 2014. Ebola Virus VP24 Targets a Unique NLS Binding Site on Karyopherin Alpha 5 to Selectively Compete with Nuclear Import of Phosphorylated STAT1. Cell Host & Microbe, 16(2), 187-200. https://doi.org/10.1016/j.chom.2014.07.008"
},
{
"DOI": "10.1016/j.antiviral.2020.104760",
"doi-asserted-by": "crossref",
"key": "75872",
"unstructured": "Yang, S. N. Y., Atkinson, S. C., Wang, C., Lee, A., Bogoyevitch, M. A., Borg, N. A. and Jans, D. A. 2020. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Research, 177, 104760. https://doi.org/10.1016/j.antiviral.2020.104760"
},
{
"DOI": "10.1016/j.bbagen.2008.07.009",
"doi-asserted-by": "crossref",
"key": "75873",
"unstructured": "Ye, Z., Wong, C. K., Li, P. and Xie, Y. 2008. A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNK-dependent apoptosis. Biochimica et Biophysica Acta (BBA) - General Subjects, 1780(12), 1383-1387. https://doi.org/10.1016/j.bbagen.2008.07.009"
},
{
"DOI": "10.1128/JVI.02371-08",
"doi-asserted-by": "crossref",
"key": "75874",
"unstructured": "Zhao, J., Falcón, A., Zhou, H., Netland, J., Enjuanes, L., Breña, P. P. and Perlman, S. 2009. Severe Acute Respiratory Syndrome Coronavirus Protein 6 Is Required for Optimal Replication. Journal of Virology, 83(5), 2368-2373. https://doi.org/10.1128/JVI.02371-08"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"doi-asserted-by": "crossref",
"key": "75875",
"unstructured": "Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., Si, H. R., Zhu, Y., Li, B., Huang, C. L., Chen, H. D., Chen, J. C., Luo, Y., Guo, H., Jiang, R. D., Liu, M. Q., Chen, Y. C., Shen, Z. R., Wang, X., Zheng, X. S., Zhao, K., Chen, Q. J., Deng, F., Liu, L. L., Yan, B., Zhan, F. X., Wang, Y. Y., Xiao, G. F. and Shi, Z. L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270-273. https://doi.org/10.1038/s41586-020-2012-7"
}
],
"reference-count": 79,
"references-count": 79,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.biotecnia.unison.mx/index.php/biotecnia/article/view/2485"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Binding affinities analysis of ivermectin, nucleocapsid and ORF6 proteins of SARS-CoV-2 to human importins α isoforms: A computational approach",
"type": "journal-article",
"volume": "27"
}