In Vitro Analysis of SARS-CoV-2 Spike Protein and Ivermectin Interaction
Alejandra García-Aguilar, Rebec Campi-Caballer, Giovani Visoso-Carvajal, José Rubén García-Sánchez, José Correa-Basurto, Jazmín García-Machorro, Judith Espinosa-Raya
doi:10.3390/ijms242216392
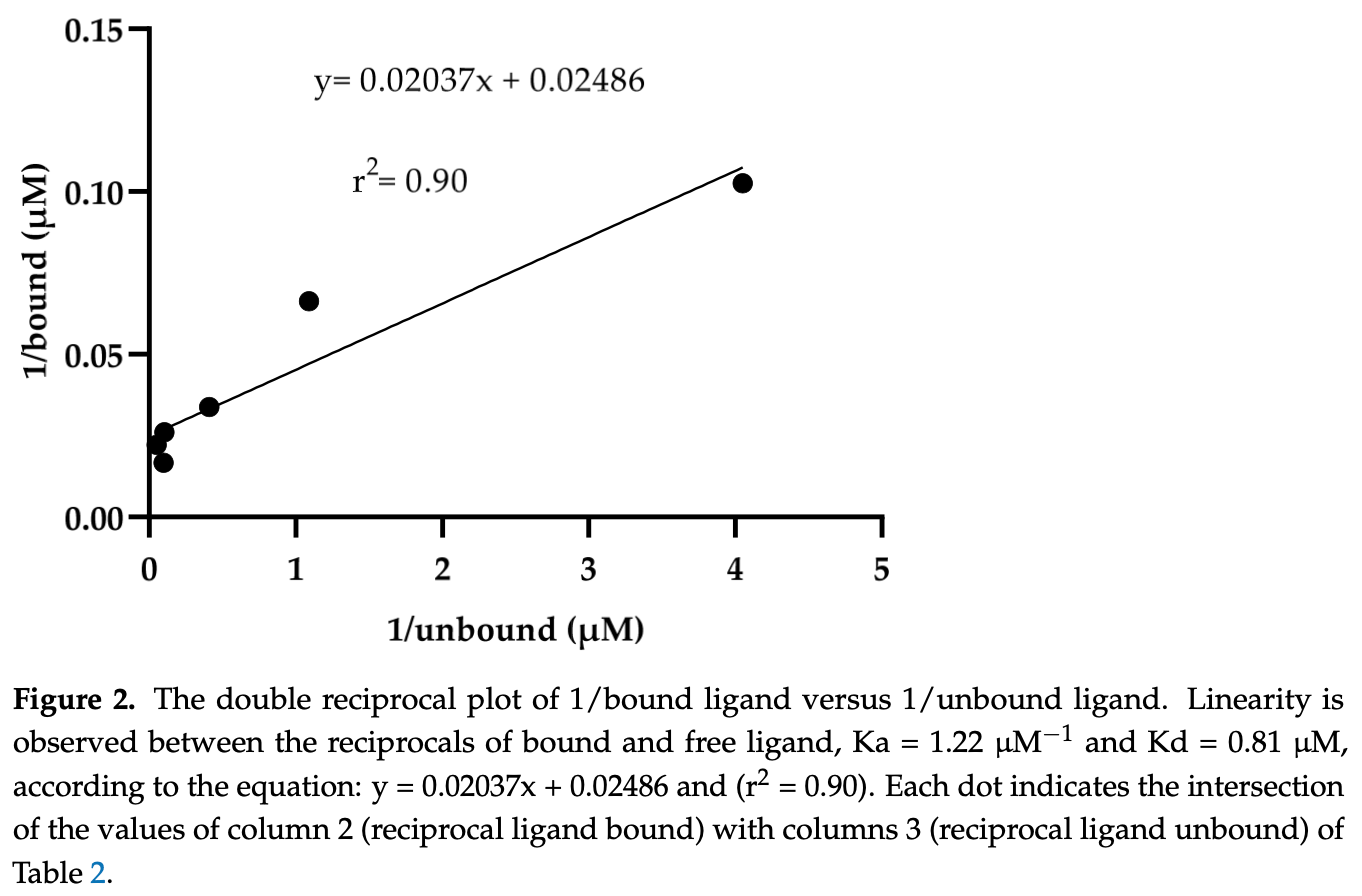
The spike (S) protein of SARS-CoV-2 is a molecular target of great interest for developing drug therapies against COVID-19 because S is responsible for the interaction of the virus with the host cell receptor. Currently, there is no outpatient safety treatment for COVID-19 disease. Furthermore, we consider it of worthy importance to evaluate experimentally the possible interaction of drugs (approved by the Food and Drug Administration) and the S, considering some previously in silico and clinical use. Then, the objective of this study was to demonstrate the in vitro interaction of ivermectin with S. The equilibrium dialysis technique with UV-Vis was performed to obtain the affinity and dissociation constants. In addition, the Drug Affinity Responsive Target Stability (DARTS) technique was used to demonstrate the in vitro interaction of S with ivermectin. The results indicate the interaction between ivermectin and the S with an association and dissociation constant of Ka = 1.22 µM -1 and Kd = 0.81 µM, respectively. The interaction was demonstrated in ratios of 1:50 pmol and 1:100 pmol (S: ivermectin) by the DARTS technique. The results obtained with these two different techniques demonstrate an interaction between S and ivermectin previously explored in silico, suggesting its clinical uses to stop the viral spread among susceptible human hosts.
Supplementary Materials: The supporting information can be downloaded at: https://www.mdpi. com/article/10.3390/ijms242216392/s1.
Conflicts of Interest: The authors declare no conflict of interest.
References
Agostini, Andres, Sims, Graham, Sheahan et al., Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease, mBio,
doi:10.1128/mBio.00221-18Awad, Hassan, Dweek, Aboelata, Rawas-Qalaji et al., Repurposing Potential of the Antiparasitic Agent Ivermectin for the Treatment and/or Prophylaxis of COVID-19, Pharmaceuticals,
doi:10.3390/ph15091068Azam, Taban, Eid, Iqbal, Alam et al., An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin alpha, J. Biomol. Struct. Dyn,
doi:10.1080/07391102.2020.1841028Bell, Ligand Binding Module
Chaccour, Casellas, Blanco-Di Matteo, Pineda, Fernandez-Montero et al., The effect of early treatment with ivermectin on viral load, symptoms and humoral response in patients with non-severe COVID-19: A pilot, double-blind, placebo-controlled, randomized clinical trial, EClinicalMedicine,
doi:10.1016/j.eclinm.2020.100720Choudhury, Das, Patra, Bhattacharya, Ghosh et al., Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: An in silico approach, Future Virol,
doi:10.2217/fvl-2020-0342Clinicaltrials, Gov, Clinical Trial of Ivermectin Plus Doxycycline for the Treatment of Confirmed Covid-19 Infection
Clinicaltrials, Gov, Efficacy, Safety and Tolerability of Ivermectin in Subjects Infected with SARS-CoV-2 with or without Symptoms (SILVERBULLET)
Clinicaltrials, Gov, Ivermectin for Severe COVID-19 Management
Correa-Basurto, Romero-Castro, Correa-Basurto, Hernandez-Rodriguez, Soriano-Ursua et al., Pharmacokinetics and tissue distribution of N-(2-hydroxyphenyl)-2-propylpentanamide in Wistar Rats and its binding properties to human serum albumin, J. Pharm. Biomed. Anal,
doi:10.1016/j.jpba.2018.09.010Deshpande, Tiwari, Nyayanit, Modak, In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2, Eur. J. Pharmacol,
doi:10.1016/j.ejphar.2020.173430Fda, Actualización Sobre El Coronavirus (COVID-19): La FDA Autoriza un Antiviral Oral Adicional
Food, Administration, Autorización de Uso de Emergencia de Paxlovid Para la Enfermedad por Coronavirus 2019
Gilead, European Commission Expands Indication for Veklury (Remdesivir) for the Treatment of Adults Not on Supplemental Oxygen and Considered High Risk for COVID-19 Disease Progression
Gonzalez, Gonzalez, Ueno, Ivermectin in Human Medicine, An Overview of the Current Status of Its Clinical Applications, Curr. Pharm. Biotechnol,
doi:10.2174/138920112800399248Gordon, Tchesnokov, Woolner, Perry, Feng et al., Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency, J. Biol. Chem,
doi:10.1074/jbc.RA120.013679Heidary, Gharebaghi, Ivermectin: A systematic review from antiviral effects to COVID-19 complementary regimen, J. Antibiot,
doi:10.1038/s41429-020-0336-zHuang, Yang, Xu, Xu, Liu, Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19, Acta Pharmacol. Sin,
doi:10.1038/s41401-020-0485-4Jans, Wagstaff, The broad spectrum host-directed agent ivermectin as an antiviral for SARS-CoV-2?, Biochem. Biophys. Res. Commun,
doi:10.1016/j.bbrc.2020.10.042Kong, Chang, Qiao, Zhao, Chen et al., Paxlovid accelerates cartilage degeneration and senescence through activating endoplasmic reticulum stress and interfering redox homeostasis, J. Transl. Med,
doi:10.1186/s12967-022-03770-4Lehrer, Rheinstein, Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2, Vivo,
doi:10.21873/invivo.12134Lomenick, Hao, Jonai, Chin, Aghajan et al., Target identification using drug affinity responsive target stability (DARTS), Proc. Natl. Acad. Sci,
doi:10.1073/pnas.0910040106Lomenick, Jung, Wohlschlegel, Huang, Target identification using drug affinity responsive target stability (DARTS), Curr. Protoc. Chem. Biol,
doi:10.1002/9780470559277.ch110180Lomenick, Olsen, Huang, Identification of direct protein targets of small molecules, ACS Chem. Biol,
doi:10.1021/cb100294vLuis, García, Unión a proteínas plasmáticas de la DL-3-hidroxi-3-etil-3-fenil-propionamida (HEPP). Un nuevo anticonvulsivante, J. Mex. Chem. Soc
Mali, Eerike, Raj, Bisoi, Priyadarshini et al., Efficacy and safety of Molnupiravir in COVID-19 patients: A systematic review, Ir. J. Med. Sci,
doi:10.1007/s11845-022-03139-yMalone, Urakova, Snijder, Campbell, Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design, Nat. Rev. Mol. Cell Biol,
doi:10.1038/s41580-021-00432-zMathachan, Sardana, Khurana, Current Use of Ivermectin in Dermatology, Tropical Medicine, and COVID-19: An Update on Pharmacology, Uses, Proven and Varied Proposed Mechanistic Action, Indian Dermatol. Online J
Merck, Merck and Ridgeback's Investigational Oral Antiviral Molnupiravir Reduced the Risk of Hospitalization or Death by Approximately 50 Percent Compared to Placebo for Patients with Mild or Moderate COVID-19 in Positive Interim Analysis of Phase 3 Study
Moctezuma, La glucoproteína spike, Rev. Mex. Mastol
Niraj, Mahajan, Prakash, Sarma, Medhi et al., A promising drug for the challenging treatment of SARS-COV-2 in the pandemic era, Indian J. Pharmacol
Pfizer, Pfizer's Novel COVID-19 Oral Antiviral Treatment Candidate Reduced Risk of Hospitalization or Death by 89% in Interim Analysis of Phase 2/3 EPIC-HR Study
Polak, Van Gool, Cohen, Von Der Thusen, Van Paassen, A systematic review of pathological findings in COVID-19: A pathophysiological timeline and possible mechanisms of disease progression, Mod. Pathol,
doi:10.1038/s41379-020-0603-3Raman, Patel, Ranjan, COVID-19: Unmasking Emerging SARS-CoV-2 Variants, Vaccines and Therapeutic Strategies, Biomolecules,
doi:10.3390/biom11070993Ramirez-Salinas, Martinez-Archundia, Correa-Basurto, Garcia-Machorro, Repositioning of Ligands That Target the Spike Glycoprotein as Potential Drugs for SARS-CoV-2 in an In Silico Study, Molecules,
doi:10.3390/molecules25235615Reina, Iglesias, Nirmatrelvir plus ritonavir (Paxlovid) a potent SARS-CoV-2 3CLpro protease inhibitor combination, Rev. Esp. Quim,
doi:10.37201/req/002.2022Sanchezruiz, Nuzum, Kouzi, Oral ivermectin for the treatment of head lice infestation, Am. J. Health-Syst. Pharm,
doi:10.2146/ajhp170464Sanderson, Hisner, Donovan-Banfield, Hartman, Lochen et al., A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes, Nature,
doi:10.1038/s41586-023-06649-6Saravolatz, Depcinski, Sharma, Molnupiravir and Nirmatrelvir-Ritonavir: Oral Coronavirus Disease 2019 Antiviral Drugs, Clin. Infect. Dis,
doi:10.1093/cid/ciac180Tang, Bidon, Jaimes, Whittaker, Daniel, Coronavirus membrane fusion mechanism offers a potential target for antiviral development, Antivir. Res,
doi:10.1016/j.antiviral.2020.104792Tregoning, Flight, Higham, Wang, Pierce, Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape, Nat. Rev. Immunol,
doi:10.1038/s41577-021-00592-1Vuignier, Schappler, Veuthey, Carrupt, Martel, Drug-protein binding: A critical review of analytical tools, Anal. Bioanal. Chem,
doi:10.1007/s00216-010-3737-1Wang, Zhao, Gao, Gao, Wang et al., SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development, Front. Cell Infect. Microbiol,
doi:10.3389/fcimb.2020.587269Wen, Yan, Sun, Fang, Yang et al., A randomized, double-blind, multicenter clinical trial on the efficacy of ivermectin against intestinal nematode infections in China, Acta Trop,
doi:10.1016/j.actatropica.2008.03.007Zhou, Wang, Liu, Lu, Dong et al., Probing antiviral drugs against SARS-CoV-2 through virus-drug association prediction based on the KATZ method, Genomics,
doi:10.1016/j.ygeno.2020.07.044