In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease
Yuri Alves De Oliveira Só, Kelton Silva Bezerra, Ricardo Gargano, Fabio L L Mendonça, Janeusa Trindade De Souto, Umberto L Fulco, Marcelo Lopes Pereira Junior, Luiz Antônio Ribeiro Júnior
doi:10.20944/preprints202404.1825.v1
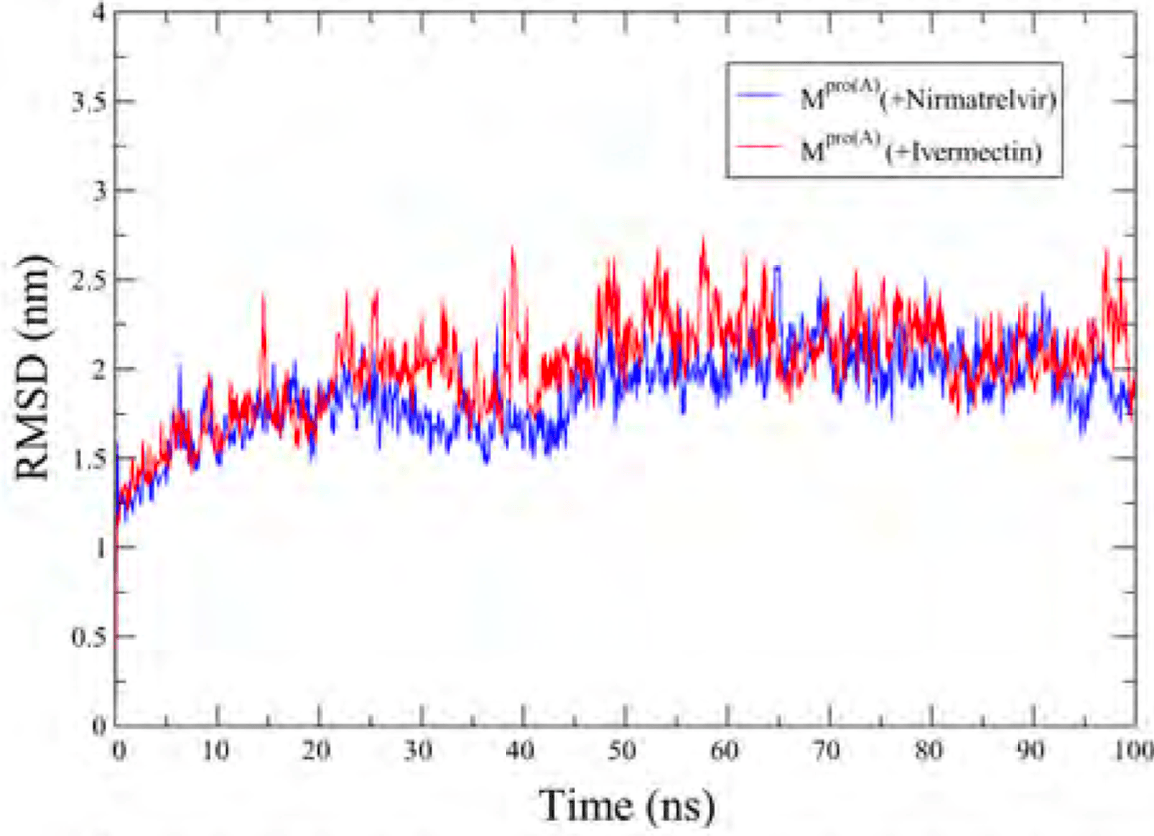
Exploring therapeutic options is crucial in the ongoing COVID-19 pandemic caused by SARS-CoV-2. Nirmatrelvir, a potent inhibitor targeting the SARS-CoV-2 M pro , shows promise as an antiviral treatment. Additionally, Ivermectin, a broad-spectrum antiparasitic drug, has demonstrated effectiveness against the virus in laboratory settings. However, its clinical implications are still debated. Using computational methods such as molecular docking and 100 ns molecular dynamics simulations, we investigated how Nirmatrelvir and Ivermectin interact with SARS-CoV-2 M pro(A) . Calculations using density functional theory have been instrumental in elucidating the behavior of isolated molecules, primarily by analyzing the frontier molecular orbitals. Our analysis revealed distinct binding patterns: Nirmatrelvir formed strong interactions with amino acids like MET49, MET165, HIS41, HIS163, HIS164, PHE140, CYS145, GLU166, and ASN142, showing stable binding with a root mean square deviation (RMSD) of around 2.0 Å. On the other hand, Ivermectin interacted with THR237, THR239, LEU271, LEU272, and LEU287, displaying an RMSD of 1.87 Å, indicating enduring interactions. Both ligands stabilized M pro (A) , with Ivermectin showing stability and persistent interactions despite forming fewer hydrogen bonds. These findings offer detailed insights into how Nirmatrelvir and Ivermectin bind to the SARS-CoV-2 main protease, providing valuable information for potential therapeutic strategies against COVID-19.
Institutional Review Board Statement: Not applicable.
Informed Consent Statement: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest.
References
Agost-Beltrán, De La Hoz-Rodríguez, Bou-Iserte, Rodríguez, Fernández-De-La Pradilla et al., Advances in the development of SARS-CoV-2 Mpro inhibitors, Molecules
Ahmad, Batool, Ain, Kim, Choi, Exploring the binding mechanism of PF-07321332 SARS-CoV-2 protease inhibitor through molecular dynamics and binding free energy simulations, International journal of molecular sciences
Alugubelli, Geng, Yang, Shaabani, Khatua et al., A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals, European Journal of Medicinal Chemistry
Andi, Kumaran, Kreitler, Soares, Keereetaweep et al., Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease, Scientific reports
Asadi, Airborne Infectious Disease Transmission Via Expiratory Aerosol Particles and Aerosolized Fomites
Atzrodt, Maknojia, Mccarthy, Oldfield, Po et al., A Guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2, The FEBS journal
Banerjee, Perera, Tillekeratne, Potential SARS-CoV-2 main protease inhibitors, Drug Discovery Today
Becke, A new mixing of Hartree-Fock and local density-functional theories, The Journal of chemical physics
Buonfrate, Chesini, Martini, Roncaglioni, Fernandez et al., High-dose ivermectin for early treatment of COVID-19 (COVER study): a randomised, double-blind, multicentre, phase II, dose-finding, proof-of-concept clinical trial, International journal of antimicrobial agents
Cannalire, Cerchia, Beccari, Di Leva, Summa, Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities, Journal of medicinal chemistry
Cobos-Campos, Apiñaniz, Parraza, Cordero, García et al., Potential use of ivermectin for the treatment and prophylaxis of SARS-CoV-2 infection, Current Research in Translational Medicine
Di Chio, Previti, Amendola, Ravichandran, Wagner et al., Development of novel dipeptide nitriles as inhibitors of rhodesain of Trypanosoma brucei rhodesiense, European Journal of Medicinal Chemistry
Frisch, Trucks, Schlegel, Scuseria, Robb et al., None, Gaussian
Fukunishi, Yamashita, Mashimo, Nakamura, Prediction of protein-compound binding energies from known activity data: docking-score-based method and its applications, Molecular Informatics
Hill, Garratt, Levi, Falconer, Ellis et al., Retracted: meta-analysis of randomized trials of ivermectin to treat SARS-CoV-2 infection. Open forum infectious diseases
Huey, Morris, Forli, Using AutoDock 4 and AutoDock vina with AutoDockTools: a tutorial
Jayaweera, Perera, Gunawardana, Manatunge, Transmission of COVID-19 virus by droplets and aerosols: A critical review on the unresolved dichotomy, Environmental research
Jin, Du, Xu, Deng, Liu et al., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature
Klauda, Venable, Freites, O'connor, Tobias et al., Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types, The journal of physical chemistry B
Lin, Zeng, Duan, Yang, Ma et al., Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants, Communications Biology
Louten, Virus transmission and epidemiology, Essential human virology
Miura, Malla, Owen, Tumber, Brewitz et al., In vitro selection of macrocyclic peptide inhibitors containing cyclic γ2, 4-amino acids targeting the SARS-CoV-2 main protease, Nature Chemistry
Parr, Density functional theory, Annual Review of Physical Chemistry
Pellis, Scarabel, Stage, Overton, Chappell et al., Challenges in control of COVID-19: short doubling time and long delay to effect of interventions, Philosophical Transactions of the Royal Society B
Rajeev, Prathiviraj, Kiran, Selvin, Zoonotic evolution and implications of microbiome in viral transmission and infection, Virus research
Sham, Schlüter, Density-functional theory of the energy gap, Physical review letters
Sharun, Dhama, Patel, Pathak, Tiwari et al., a new candidate therapeutic against SARS-CoV-2/COVID-19
Stewart, Application of the PM6 method to modeling proteins, Journal of molecular modeling
Tyndall, S-217622, a 3CL protease inhibitor and clinical candidate for SARS-CoV-2, Journal of Medicinal Chemistry
Weigend, Ahlrichs, Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy, Physical Chemistry Chemical Physics
Weiss, Sankaran, Emergence of epidemic diseases: Zoonoses and other origins, Faculty Reviews
Worldometer, COVID-19 CORONAVIRUS PANDEMIC
Yamamoto, Yasuo, Sekijima, Screening for inhibitors of main protease in sars-cov-2: In silico and in vitro approach avoiding peptidyl secondary amides, Journal of Chemical Information and Modeling
Zaidi, Dehgani-Mobaraki, The mechanisms of action of ivermectin against SARS-CoV-2-an extensive review, The Journal of antibiotics
Zhao, Fang, Zhang, Zhang, Zhao et al., Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332, Protein & cell
DOI record:
{
"DOI": "10.20944/preprints202404.1825.v1",
"URL": "http://dx.doi.org/10.20944/preprints202404.1825.v1",
"abstract": "<jats:p>Exploring therapeutic options is crucial in the ongoing COVID-19 pandemic caused by SARS-CoV-2. Nirmatrelvir, a potent inhibitor targeting the SARS-CoV-2 Mpro, shows promise as an antiviral treatment. Additionally, Ivermectin, a broad-spectrum antiparasitic drug, has demonstrated effectiveness against the virus in laboratory settings. However, its clinical implications are still debated. Using computational methods such as molecular docking and 100 ns molecular dynamics simulations, we investigated how Nirmatrelvir and Ivermectin interact with SARS-CoV-2 Mpro(A). Calculations using density functional theory have been instrumental in elucidating the behavior of isolated molecules, primarily by analyzing the frontier molecular orbitals. Our analysis revealed distinct binding patterns: Nirmatrelvir formed strong interactions with amino acids like MET49, MET165, HIS41, HIS163, HIS164, PHE140, CYS145, GLU166, and ASN142, showing stable binding with a root mean square deviation (RMSD) of around 2.0 Å. On the other hand, Ivermectin interacted with THR237, THR239, LEU271, LEU272, and LEU287, displaying an RMSD of 1.87 Å, indicating enduring interactions. Both ligands stabilized Mpro(A), with Ivermectin showing stability and persistent interactions despite forming fewer hydrogen bonds. These findings offer detailed insights into how Nirmatrelvir and Ivermectin bind to the SARS-CoV-2 main protease, providing valuable information for potential therapeutic strategies against COVID-19.</jats:p>",
"accepted": {
"date-parts": [
[
2024,
4,
26
]
]
},
"author": [
{
"affiliation": [],
"family": "de Oliveira Só",
"given": "Yuri Alves",
"sequence": "first"
},
{
"affiliation": [],
"family": "Bezerra",
"given": "Kelton Silva",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-3823-7436",
"affiliation": [],
"authenticated-orcid": false,
"family": "Gargano",
"given": "Ricardo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mendonça",
"given": "Fabio L.L",
"sequence": "additional"
},
{
"affiliation": [],
"family": "de Souto",
"given": "Janeusa Trindade",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Fulco",
"given": "Umberto L.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pereira Junior",
"given": "Marcelo Lopes",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Júnior",
"given": "Luiz Antônio Ribeiro",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
4,
29
]
],
"date-time": "2024-04-29T08:42:45Z",
"timestamp": 1714380165000
},
"deposited": {
"date-parts": [
[
2024,
4,
29
]
],
"date-time": "2024-04-29T08:48:59Z",
"timestamp": 1714380539000
},
"group-title": "Biology and Life Sciences",
"indexed": {
"date-parts": [
[
2024,
4,
30
]
],
"date-time": "2024-04-30T00:29:13Z",
"timestamp": 1714436953418
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
4,
28
]
]
},
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
4,
28
]
],
"date-time": "2024-04-28T00:00:00Z",
"timestamp": 1714262400000
}
}
],
"member": "1968",
"original-title": [],
"posted": {
"date-parts": [
[
2024,
4,
28
]
]
},
"prefix": "10.20944",
"published": {
"date-parts": [
[
2024,
4,
28
]
]
},
"publisher": "MDPI AG",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.preprints.org/manuscript/202404.1825/v1"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease",
"type": "posted-content"
}