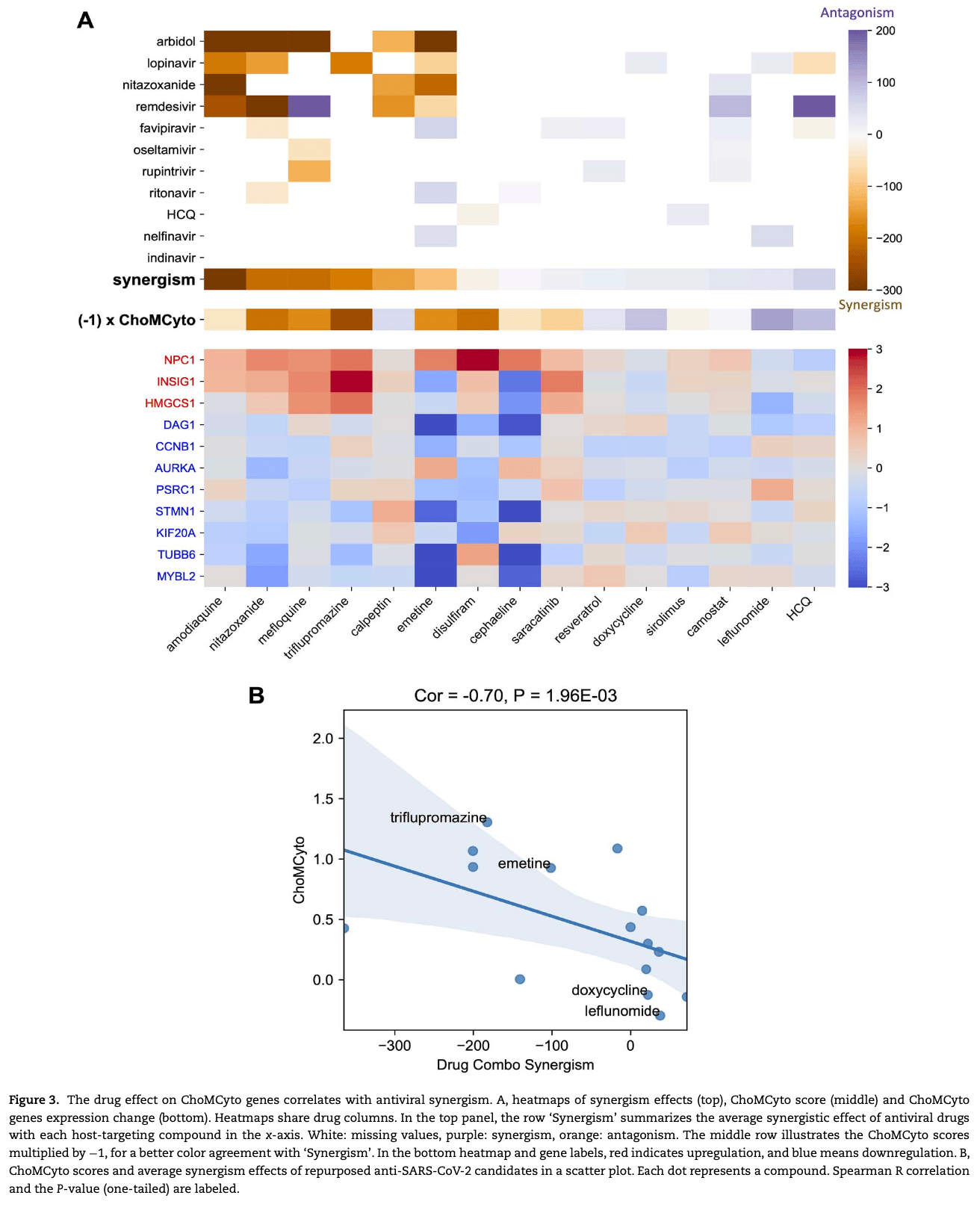
Published anti-SARS-CoV-2 in vitro hits share common mechanisms of action that synergize with antivirals
Jing Xing, Shreya Paithankar, Ke Liu, Katie Uhl, Xiaopeng Li, Meehyun Ko, Seungtaek Kim, Jeremy Haskins, Bin Chen
Briefings in Bioinformatics, doi:10.1093/bib/bbab249
The global efforts in the past year have led to the discovery of nearly 200 drug repurposing candidates for COVID-19. Gaining more insights into their mechanisms of action could facilitate a better understanding of infection and the development of therapeutics. Leveraging large-scale drug-induced gene expression profiles, we found 36% of the active compounds regulate genes related to cholesterol homeostasis and microtubule cytoskeleton organization. Following bioinformatics analyses revealed that the expression of these genes is associated with COVID-19 patient severity and has predictive power on anti-SARS-CoV-2 efficacy in vitro. Monensin, a top new compound that regulates these genes, was further confirmed as an inhibitor of SARS-CoV-2 replication in Vero-E6 cells. Interestingly, drugs co-targeting cholesterol homeostasis and microtubule cytoskeleton organization processes more likely present a synergistic effect with antivirals. Therefore, potential therapeutics could be centered around combinations of targeting these processes and viral proteins.
Supplementary Data Supplementary data are available online at Briefings in Bioinformatics.
Contributions J.X. and B.C. conceived the study. J.X. led and performed all the analyses with the input from S.P., K.L., J.H., and B.C., K.U. and X.L. performed the experiments in human lung primary small airway cells. M.K. and S.K. performed in vitro efficacy studies. J.X. and B.C. wrote the manuscript with the input from all co-authors. B.C. supervised the study.
References
Araki, Ebata, Guo, p53 regulates cytoskeleton remodeling to suppress tumor progression, Cell Mol Life Sci
Aylon, The hippo pathway, p53 and cholesterol, Cell Cycle
Bobrowski, Chen, Eastman, Synergistic and antagonistic drug combinations against SARS-CoV-2, Mol Ther
Chen, Greenside, Paik, Relating chemical structure to cellular response: an integrative analysis of gene expression, bioactivity, and structural data across 11,000 compounds, CPT Pharmacometrics Syst Pharmacol
Chen, Ma, Paik, Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets, Nat Commun
Chen, Wei, Ma, Computational discovery of niclosamide ethanolamine, a repurposed drug candidate that reduces growth of hepatocellular carcinoma cells in vitro and in mice by inhibiting cell division cycle 37 signaling, Gastroenterology
Cortese, Lee, Cerikan, Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies, Cell Host Microbe
Daniloski, Jordan, Wessels, Identification of required host factors for SARS-CoV-2 infection in human cells, Cell
Dobin, Davis, Schlesinger, STAR: ultrafast universal RNA-seq aligner, Bioinformatics
Gordon, Jang, Bouhaddou, A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
Griffiths, Quinn, Warren, Dissection of the Golgi complex. I. Monensin inhibits the transport of viral membrane proteins from medial to trans Golgi cisternae in baby hamster kidney cells infected with Semliki Forest virus, J Cell Biol
Hoffmann, Schneider, Rozen-Gagnon, TMEM41B is a pan-flavivirus host factor, Cell
Jeon, Ko, Lee, Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs, Antimicrob Agents Chemother
Lee, Tiong, Chang, DeSigN: connecting gene expression with therapeutics for drug repurposing and development, BMC Genomics
Li, Dewey, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome, BMC Bioinformatics
Li, Ruotti, Stewart, RNA-Seq gene expression estimation with read mapping uncertainty, Bioinformatics
Li, Tang, Buonfiglio, Electrolyte transport properties in distal small airways from cystic fibrosis pigs with implications for host defense, Am J Physiol Lung Cell Mol Physiol
Luo, Yang, Song, Mechanisms and regulation of cholesterol homeostasis, Nat Rev Mol Cell Biol
Lyu, Yang, Head, Pharmacological blockade of cholesterol trafficking by cepharanthine in endothelial cells suppresses angiogenesis and tumor growth, Cancer Lett
Mendez, Gaulton, Bento, ChEMBL: towards direct deposition of bioassay data, Nucleic Acids Res
Mirabelli, Wotring, Zhang, Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19, bioRxiv,
doi:10.1101/2020.05.27.117184Riva, Yuan, Yin, Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing, Nature
Schneider, Luna, Hoffmann, Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks, Cell
Subramanian, Narayan, Corsello, A next generation connectivity map: L1000 platform and the first 1,000,000 profiles, Cell
Szklarczyk, Gable, Lyon, STRING v11: proteinprotein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets, Nucleic Acids Res
Van Noort, Schölch, Iskar, Novel drug candidates for the treatment of metastatic colorectal cancer through global inverse gene-expression profiling, Cancer Res
Wang, Simoneau, Kulsuptrakul, Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses, Cell
Wei, Alfajaro, Deweirdt, Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection, Cell
Wei, Zeng, Su, Hypolipidemia is associated with the severity of COVID-19, J Clin Lipidol
White, Rosales, Yildiz, Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A, Science
Xing, Shankar, Drelich, Analysis of infected host gene expression reveals repurposed drug candidates and time-dependent host response dynamics for COVID-19,
doi:10.1101/2020.04.07.030734Zang, Case, Yutuc, Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion, Proc Natl Acad Sci U S A
Zeng, Glicksberg, Newbury, OCTAD: an open workspace for virtually screening therapeutics targeting precise cancer patient groups using gene expression features, Nat Protoc
Zu, Deng, Zhou, 25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor, Cell Res
DOI record:
{
"DOI": "10.1093/bib/bbab249",
"ISSN": [
"1467-5463",
"1477-4054"
],
"URL": "http://dx.doi.org/10.1093/bib/bbab249",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>The global efforts in the past year have led to the discovery of nearly 200 drug repurposing candidates for COVID-19. Gaining more insights into their mechanisms of action could facilitate a better understanding of infection and the development of therapeutics. Leveraging large-scale drug-induced gene expression profiles, we found 36% of the active compounds regulate genes related to cholesterol homeostasis and microtubule cytoskeleton organization. Following bioinformatics analyses revealed that the expression of these genes is associated with COVID-19 patient severity and has predictive power on anti-SARS-CoV-2 efficacy in vitro. Monensin, a top new compound that regulates these genes, was further confirmed as an inhibitor of SARS-CoV-2 replication in Vero-E6 cells. Interestingly, drugs co-targeting cholesterol homeostasis and microtubule cytoskeleton organization processes more likely present a synergistic effect with antivirals. Therefore, potential therapeutics could be centered around combinations of targeting these processes and viral proteins.</jats:p>",
"author": [
{
"ORCID": "http://orcid.org/0000-0003-4726-8525",
"affiliation": [
{
"name": "Department of Pediatrics and Human Development , , Grand Rapids, Michigan, USA"
},
{
"name": "Michigan State University , , Grand Rapids, Michigan, USA"
}
],
"authenticated-orcid": false,
"family": "Xing",
"given": "Jing",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Pediatrics and Human Development , , Grand Rapids, Michigan, USA"
},
{
"name": "Michigan State University , , Grand Rapids, Michigan, USA"
}
],
"family": "Paithankar",
"given": "Shreya",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-3190-1656",
"affiliation": [
{
"name": "Department of Pediatrics and Human Development , , Grand Rapids, Michigan, USA"
},
{
"name": "Michigan State University , , Grand Rapids, Michigan, USA"
}
],
"authenticated-orcid": false,
"family": "Liu",
"given": "Ke",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics and Human Development , , Grand Rapids, Michigan, USA"
},
{
"name": "Michigan State University , , Grand Rapids, Michigan, USA"
}
],
"family": "Uhl",
"given": "Katie",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics and Human Development , , Grand Rapids, Michigan, USA"
},
{
"name": "Michigan State University , , Grand Rapids, Michigan, USA"
}
],
"family": "Li",
"given": "Xiaopeng",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Zoonotic Virus Laboratory, Institut Pasteur Korea , Seongnam, South Korea"
}
],
"family": "Ko",
"given": "Meehyun",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Zoonotic Virus Laboratory, Institut Pasteur Korea , Seongnam, South Korea"
}
],
"family": "Kim",
"given": "Seungtaek",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics and Human Development , , Grand Rapids, Michigan, USA"
},
{
"name": "Michigan State University , , Grand Rapids, Michigan, USA"
}
],
"family": "Haskins",
"given": "Jeremy",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics and Human Development , , Grand Rapids, Michigan, USA"
},
{
"name": "Michigan State University , , Grand Rapids, Michigan, USA"
},
{
"name": "Department of Pharmacology and Toxicology , , Grand Rapids, Michigan, USA"
},
{
"name": "Michigan State University , , Grand Rapids, Michigan, USA"
}
],
"family": "Chen",
"given": "Bin",
"sequence": "additional"
}
],
"container-title": "Briefings in Bioinformatics",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
7,
6
]
],
"date-time": "2021-07-06T19:11:13Z",
"timestamp": 1625598673000
},
"deposited": {
"date-parts": [
[
2022,
7,
9
]
],
"date-time": "2022-07-09T00:07:37Z",
"timestamp": 1657325257000
},
"funder": [
{
"DOI": "10.13039/100000002",
"award": [
"R01GM134307",
"K01ES028047"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100000002",
"id-type": "DOI"
}
],
"name": "National Institutes of Health"
},
{
"DOI": "10.13039/100000001",
"award": [
"2028717"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100000001",
"id-type": "DOI"
}
],
"name": "National Science Foundation"
},
{
"award": [
"NCCP43326"
],
"name": "National Culture Collection for Pathogens"
}
],
"indexed": {
"date-parts": [
[
2024,
8,
27
]
],
"date-time": "2024-08-27T07:16:21Z",
"timestamp": 1724742981584
},
"is-referenced-by-count": 8,
"issue": "6",
"issued": {
"date-parts": [
[
2021,
7,
9
]
]
},
"journal-issue": {
"issue": "6",
"published-print": {
"date-parts": [
[
2021,
11,
5
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://academic.oup.com/journals/pages/open_access/funder_policies/chorus/standard_publication_model",
"content-version": "vor",
"delay-in-days": 1,
"start": {
"date-parts": [
[
2021,
7,
10
]
],
"date-time": "2021-07-10T00:00:00Z",
"timestamp": 1625875200000
}
}
],
"link": [
{
"URL": "https://academic.oup.com/bib/article-pdf/22/6/bbab249/41088851/bbab249.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "syndication"
},
{
"URL": "https://academic.oup.com/bib/article-pdf/22/6/bbab249/41088851/bbab249.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "286",
"original-title": [],
"prefix": "10.1093",
"published": {
"date-parts": [
[
2021,
7,
9
]
]
},
"published-online": {
"date-parts": [
[
2021,
7,
9
]
]
},
"published-other": {
"date-parts": [
[
2021,
11
]
]
},
"published-print": {
"date-parts": [
[
2021,
11,
5
]
]
},
"publisher": "Oxford University Press (OUP)",
"reference": [
{
"DOI": "10.1128/AAC.00819-20",
"article-title": "Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs",
"author": "Jeon",
"doi-asserted-by": "crossref",
"first-page": "e00819",
"journal-title": "Antimicrob Agents Chemother",
"key": "2022070900070061800_ref1",
"volume": "64",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2577-1",
"article-title": "Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing",
"author": "Riva",
"doi-asserted-by": "crossref",
"first-page": "113",
"journal-title": "Nature",
"key": "2022070900070061800_ref2",
"volume": "586",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"article-title": "A SARS-CoV-2 protein interaction map reveals targets for drug repurposing",
"author": "Gordon",
"doi-asserted-by": "crossref",
"first-page": "459",
"journal-title": "Nature",
"key": "2022070900070061800_ref3",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1126/science.abf4058",
"article-title": "Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A",
"author": "White",
"doi-asserted-by": "crossref",
"first-page": "926",
"journal-title": "Science",
"key": "2022070900070061800_ref4",
"volume": "371",
"year": "2021"
},
{
"DOI": "10.1186/s12864-016-3260-7",
"article-title": "DeSigN: connecting gene expression with therapeutics for drug repurposing and development",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "934",
"journal-title": "BMC Genomics",
"key": "2022070900070061800_ref5",
"volume": "18",
"year": "2017"
},
{
"DOI": "10.1158/0008-5472.CAN-13-3540",
"article-title": "Novel drug candidates for the treatment of metastatic colorectal cancer through global inverse gene-expression profiling",
"author": "Noort",
"doi-asserted-by": "crossref",
"first-page": "5690",
"journal-title": "Cancer Res",
"key": "2022070900070061800_ref6",
"volume": "74",
"year": "2014"
},
{
"DOI": "10.1038/ncomms16022",
"article-title": "Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "16022",
"journal-title": "Nat Commun",
"key": "2022070900070061800_ref7",
"volume": "8",
"year": "2017"
},
{
"DOI": "10.1053/j.gastro.2017.02.039",
"article-title": "Computational discovery of niclosamide ethanolamine, a repurposed drug candidate that reduces growth of hepatocellular carcinoma cells in vitro and in mice by inhibiting cell division cycle 37 signaling",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "2022",
"journal-title": "Gastroenterology",
"key": "2022070900070061800_ref8",
"volume": "152",
"year": "2017"
},
{
"DOI": "10.1016/j.cell.2017.10.049",
"article-title": "A next generation connectivity map: L1000 platform and the first 1,000,000 profiles",
"author": "Subramanian",
"doi-asserted-by": "crossref",
"first-page": "1437",
"journal-title": "Cell",
"key": "2022070900070061800_ref9",
"volume": "171",
"year": "2017"
},
{
"DOI": "10.1016/j.cell.2020.12.005",
"article-title": "TMEM41B is a pan-flavivirus host factor",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "133",
"journal-title": "Cell",
"key": "2022070900070061800_ref10",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.12.006",
"article-title": "Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks",
"author": "Schneider",
"doi-asserted-by": "crossref",
"first-page": "120",
"journal-title": "Cell",
"key": "2022070900070061800_ref11",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.12.004",
"article-title": "Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "106",
"journal-title": "Cell",
"key": "2022070900070061800_ref12",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"article-title": "Identification of required host factors for SARS-CoV-2 infection in human cells",
"author": "Daniloski",
"doi-asserted-by": "crossref",
"first-page": "92",
"journal-title": "Cell",
"key": "2022070900070061800_ref13",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.10.028",
"article-title": "Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection",
"author": "Wei",
"doi-asserted-by": "crossref",
"first-page": "76",
"journal-title": "Cell",
"key": "2022070900070061800_ref14",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41580-019-0190-7",
"article-title": "Mechanisms and regulation of cholesterol homeostasis",
"author": "Luo",
"doi-asserted-by": "crossref",
"first-page": "225",
"journal-title": "Nat Rev Mol Cell Biol",
"key": "2022070900070061800_ref15",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2020.11.003",
"article-title": "Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies",
"author": "Cortese",
"doi-asserted-by": "crossref",
"first-page": "853",
"journal-title": "Cell Host Microbe",
"key": "2022070900070061800_ref16",
"volume": "28",
"year": "2020"
},
{
"DOI": "10.1016/j.canlet.2017.09.009",
"article-title": "Pharmacological blockade of cholesterol trafficking by cepharanthine in endothelial cells suppresses angiogenesis and tumor growth",
"author": "Lyu",
"doi-asserted-by": "crossref",
"first-page": "91",
"journal-title": "Cancer Lett",
"key": "2022070900070061800_ref17",
"volume": "409",
"year": "2017"
},
{
"article-title": "Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19",
"author": "Mirabelli",
"journal-title": "bioRxiv",
"key": "2022070900070061800_ref18",
"year": "2020"
},
{
"DOI": "10.1101/2020.06.04.135046",
"article-title": "An OpenData portal to share COVID-19 drug repurposing data in real time",
"author": "Brimacombe",
"doi-asserted-by": "publisher",
"journal-title": "bioRxiv",
"key": "2022070900070061800_ref19",
"year": "2020"
},
{
"DOI": "10.1016/j.ymthe.2020.12.016",
"article-title": "Synergistic and antagonistic drug combinations against SARS-CoV-2",
"author": "Bobrowski",
"doi-asserted-by": "crossref",
"first-page": "873",
"journal-title": "Mol Ther",
"key": "2022070900070061800_ref20",
"volume": "29",
"year": "2021"
},
{
"DOI": "10.1080/15384101.2016.1207840",
"article-title": "The hippo pathway, p53 and cholesterol",
"author": "Aylon",
"doi-asserted-by": "crossref",
"first-page": "2248",
"journal-title": "Cell Cycle",
"key": "2022070900070061800_ref21",
"volume": "15",
"year": "2016"
},
{
"DOI": "10.1007/s00018-015-1989-9",
"article-title": "p53 regulates cytoskeleton remodeling to suppress tumor progression",
"author": "Araki",
"doi-asserted-by": "crossref",
"first-page": "4077",
"journal-title": "Cell Mol Life Sci",
"key": "2022070900070061800_ref22",
"volume": "72",
"year": "2015"
},
{
"DOI": "10.1083/jcb.96.3.835",
"article-title": "Dissection of the Golgi complex. I. Monensin inhibits the transport of viral membrane proteins from medial to trans Golgi cisternae in baby hamster kidney cells infected with Semliki Forest virus",
"author": "Griffiths",
"doi-asserted-by": "crossref",
"first-page": "835",
"journal-title": "J Cell Biol",
"key": "2022070900070061800_ref23",
"volume": "96",
"year": "1983"
},
{
"DOI": "10.1016/j.jacl.2020.04.008",
"article-title": "Hypolipidemia is associated with the severity of COVID-19",
"author": "Wei",
"doi-asserted-by": "crossref",
"first-page": "297",
"journal-title": "J Clin Lipidol",
"key": "2022070900070061800_ref24",
"volume": "14",
"year": "2020"
},
{
"DOI": "10.1073/pnas.2012197117",
"article-title": "Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion",
"author": "Zang",
"doi-asserted-by": "crossref",
"first-page": "32105",
"journal-title": "Proc Natl Acad Sci U S A",
"key": "2022070900070061800_ref25",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.1038/s41422-020-00398-1",
"article-title": "25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor",
"author": "Zu",
"doi-asserted-by": "crossref",
"first-page": "1043",
"issue": "11",
"journal-title": "Cell Res",
"key": "2022070900070061800_ref26",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1101/2020.04.07.030734",
"article-title": "Analysis of infected host gene expression reveals repurposed drug candidates and time-dependent host response dynamics for COVID-19",
"author": "Xing",
"doi-asserted-by": "publisher",
"journal-title": "bioRxiv",
"key": "2022070900070061800_ref27",
"year": "2020"
},
{
"DOI": "10.1038/s41596-020-00430-z",
"article-title": "OCTAD: an open workspace for virtually screening therapeutics targeting precise cancer patient groups using gene expression features",
"author": "Zeng",
"doi-asserted-by": "crossref",
"first-page": "728",
"journal-title": "Nat Protoc",
"key": "2022070900070061800_ref28",
"volume": "16",
"year": "2021"
},
{
"DOI": "10.1002/psp4.12009",
"article-title": "Relating chemical structure to cellular response: an integrative analysis of gene expression, bioactivity, and structural data across 11,000 compounds",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "576",
"journal-title": "CPT Pharmacometrics Syst Pharmacol",
"key": "2022070900070061800_ref29",
"volume": "4",
"year": "2015"
},
{
"DOI": "10.1093/nar/gky1131",
"article-title": "STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets",
"author": "Szklarczyk",
"doi-asserted-by": "crossref",
"first-page": "D607",
"journal-title": "Nucleic Acids Res",
"key": "2022070900070061800_ref30",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1152/ajplung.00422.2015",
"article-title": "Electrolyte transport properties in distal small airways from cystic fibrosis pigs with implications for host defense",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "L670",
"journal-title": "Am J Physiol Lung Cell Mol Physiol",
"key": "2022070900070061800_ref31",
"volume": "310",
"year": "2016"
},
{
"DOI": "10.1093/bioinformatics/btp692",
"article-title": "RNA-Seq gene expression estimation with read mapping uncertainty",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "493",
"journal-title": "Bioinformatics",
"key": "2022070900070061800_ref32",
"volume": "26",
"year": "2010"
},
{
"DOI": "10.1186/1471-2105-12-323",
"article-title": "RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "323",
"journal-title": "BMC Bioinformatics",
"key": "2022070900070061800_ref33",
"volume": "12",
"year": "2011"
},
{
"DOI": "10.1093/bioinformatics/bts635",
"article-title": "STAR: ultrafast universal RNA-seq aligner",
"author": "Dobin",
"doi-asserted-by": "crossref",
"first-page": "15",
"journal-title": "Bioinformatics",
"key": "2022070900070061800_ref34",
"volume": "29",
"year": "2013"
},
{
"DOI": "10.1093/nar/gky1075",
"article-title": "ChEMBL: towards direct deposition of bioassay data",
"author": "Mendez",
"doi-asserted-by": "crossref",
"first-page": "D930",
"journal-title": "Nucleic Acids Res",
"key": "2022070900070061800_ref35",
"volume": "47",
"year": "2019"
}
],
"reference-count": 35,
"references-count": 35,
"relation": {
"has-preprint": [
{
"asserted-by": "object",
"id": "10.1101/2021.03.04.433931",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "https://academic.oup.com/bib/article/doi/10.1093/bib/bbab249/6318177"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Published anti-SARS-CoV-2 <i>in vitro</i> hits share common mechanisms of action that synergize with antivirals",
"type": "journal-article",
"volume": "22"
}