The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism
Professor Brian J Willett, Joe Grove, Oscar A Maclean, Craig Wilkie, Nicola Logan, Giuditta De Lorenzo, Wilhelm Furnon, Sam Scott, Maria Manali, Agnieszka Szemiel, Shirin Ashraf, Elen Vink, William T Harvey, Chris Davis, Richard Orton, Joseph Hughes, Poppy Holland, Vanessa Silva, David Pascall, Kathryn Puxty, Ana Da Silva Filipe, Gonzalo Yebra, Sharif Shaaban, Matthew T G Holden, Rute Maria Pinto, Prof Rory N Gunson, Dr Kate E Templeton, Pablo R Murcia, Arvind H Patel, John Haughney, Prof David L Robertson, Massimo Palmarini, Surajit Ray, Professor Emma C Thomson
doi:10.1101/2022.01.03.21268111
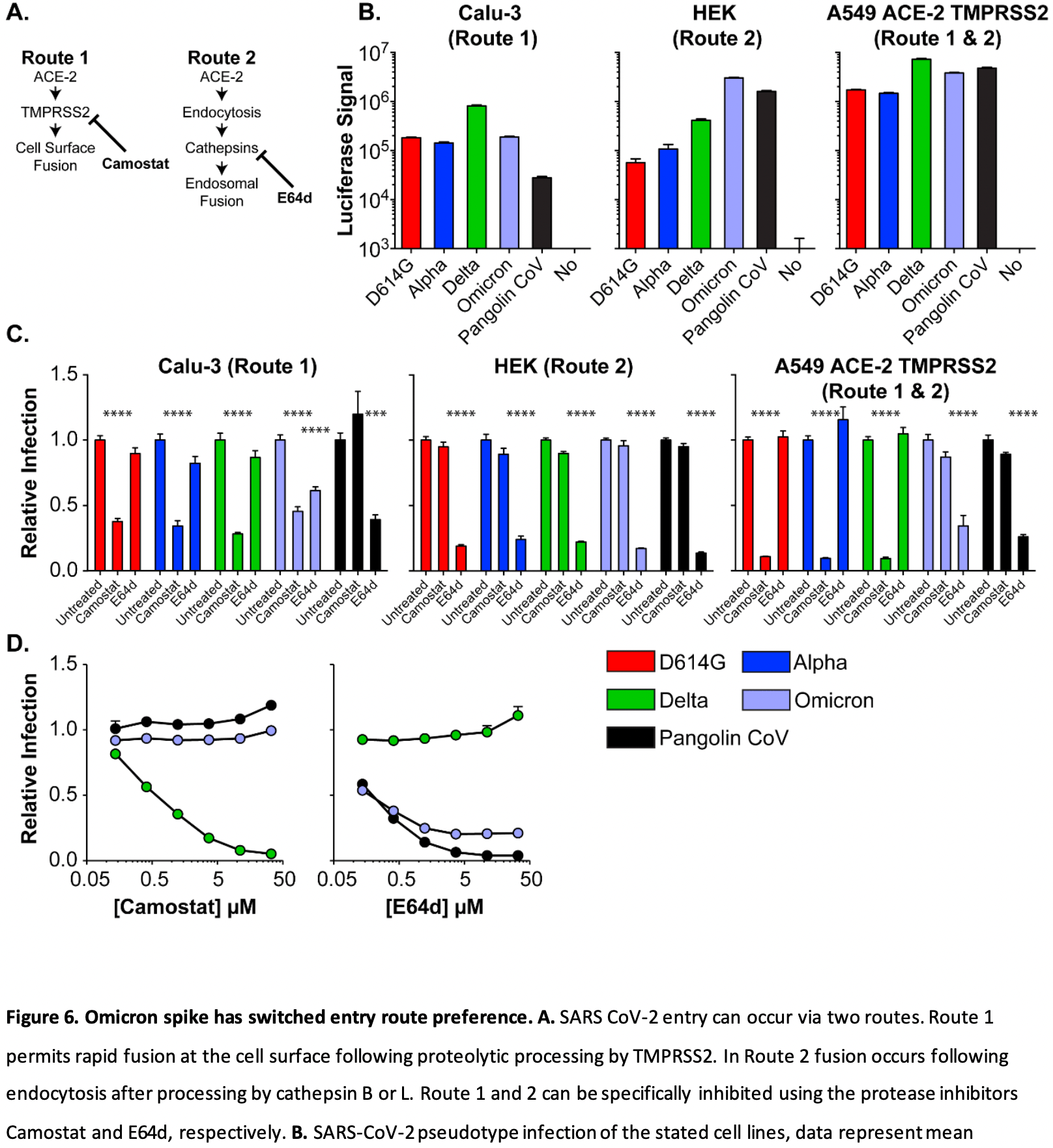
Vaccination-based exposure to spike protein derived from early SARS-CoV-2 sequences is the key public health strategy against COVID-19. Successive waves of SARS-CoV-2 infections have been characterised by the evolution of highly mutated variants that are more transmissible and that partially evade the adaptive immune response. Omicron is the fifth of these "Variants of Concern" (VOC) and is characterised by a step change in transmission capability, suggesting significant antigenic and biological change. It is characterised by 45 amino acid substitutions, including 30 changes in the spike protein relative to one of the earliest sequences, Wuhan-Hu-1, of which 15 occur in the receptorbinding domain, an area strongly associated with humoral immune evasion. In this study, we demonstrate both markedly decreased neutralisation in serology assays and real-world vaccine effectiveness in recipients of two doses of vaccine, with efficacy partially recovered by a third mRNA booster dose. We also show that immunity from natural infection (without vaccination) is more protective than two doses of vaccine but inferior to three doses. Finally, we demonstrate fundamental changes in the Omicron entry process in vitro, towards TMPRSS2-independent fusion, representing a major shift in the replication properties of SARS-CoV-2. Overall, these findings underlie rapid global transmission and may alter the clinical severity of disease associated with the Omicron variant.
Supplementary Information
Epidemiological description of the emergence of the Omicron variant in the UK On the 27 th November 2021, the UK Health Security Agency detected 2 cases of Omicron in England, the following day 6 Scottish cases were detected by community (Pillar 2) sequencing. Over the next 10 days (to 8 th December 2021) a further 95 genome sequences were obtained. Due to the rapid spread of Omicron and low genetic diversity, the genome sequences are highly related with mean genetic divergence of 1 single nucleotide polymorphisms (SNPs) and maximum 7 SNPs. The phylogenetic relationship to Omicron sequences from other countries is consistent with multiple introductions associated with travel to South Africa followed by community transmissions within Scotland. Amongst the Scottish samples diverged from the tree backbone, there were a number identified that are genetically divergent, i.e., greater than 2 single nucleotide polymorphisms from the nearest Scottish sample (Figure 1D ). Moreover, comparison to the wider international collection of Omicron samples revealed that they were more closely related to genomes from other countries than other Scottish samples. These samples therefore likely represent independent introductions to Scotland, but without more detailed epidemiological data, the number of introductions is unknown. Where there are indistinguishable samples in the phylogeny from Scotland and elsewhere in world, importation cannot be ruled out as a source..
References
Aggarwal, SARS-CoV-2 Omicron: reduction of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern, medRxiv
Ahmed, Quadeer, Mckay, SARS-CoV-2 T cell responses are expected to remain robust against Omicron
Andrews, Effectiveness of COVID-19 vaccines against the Omicron
Basile, Improved neutralization of the SARS-CoV-2 Omicron variant after Pfizer-BioNTech BNT162b2 COVID-19 vaccine boosting
Belouzard, Chu, Whittaker, Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites, Proceedings of the National Academy of Sciences
Benvenuto, Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy, Journal of Infection
Braga, Drugs that inhibit TMEM16 proteins block SARS-CoV-2 Spike-induced syncytia, Nature
Burnett, Immunizations with diverse sarbecovirus receptor-binding domains elicit SARS-CoV-2 neutralizing antibodies against a conserved site of vulnerability, Immunity
Cameroni, Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift
Cao, 1. 529 escapes the majority of SARS-CoV-2 neutralizing antibodies of diverse epitopes
Cele, SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection
Cho, Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination, Nature
Cromer, Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis, The Lancet Microbe
Daniloski, Identification of required host factors for SARS-CoV-2 infection in human cells, Cell
Davis, Reduced neutralisation of the Delta (B. 1.617. 2) SARS-CoV-2 variant of concern following vaccination, PLoS pathogens
Dejnirattisai, Reduced neutralisation of SARS-CoV-2 omicron B. 1.1. 529 variant by post-immunisation serum, The Lancet
Dicken, Characterisation of B. 1.1. 7 and Pangolin coronavirus spike provides insights on the evolutionary trajectory of SARS-CoV-2
Doria-Rose, Booster of mRNA-1273 vaccine reduces SARS-CoV-2 Omicron escape from neutralizing antibodies, medRxiv
Earle, Evidence for antibody as a protective correlate for COVID-19 vaccines, Vaccine
Feikin, Duration of Effectiveness of Vaccines Against SARS-CoV-2 Infection and COVID-19 Disease: Results of a Systematic Review and Meta-Regression, SSRN Electronic Journal,
doi:10.2139/SSRN.3961378Garcia-Beltran, mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant, Cell
Gilbert, Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial, Science
Greaney, Comprehensive mapping of mutations in the SARS-CoV-2 receptorbinding domain that affect recognition by polyclonal human plasma antibodies, Cell host & microbe
Greaney, Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies, Nature Communications
Gu, Probable Transmission of SARS-CoV-2 Omicron Variant in Quarantine Hotel, Hong Kong, China, Emerging infectious diseases
Hoffmann, Kleine-Weber, Pöhlmann, A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells, Molecular cell
Hoffmann, SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, cell
Hughes, SARS-CoV-2 serosurveillance in a patient population reveals differences in virus exposure and antibody-mediated immunity according to host demography and healthcare setting, Journal of Infectious Diseases
Jackson, Farzan, Chen, Choe, Mechanisms of SARS-CoV-2 entry into cells, Nature Reviews Molecular Cell Biology
Khoury, Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection, Nature medicine
Kim, Gordon, Sheehan, Rothberg, Duration of SARS-CoV-2 Natural Immunity and Protection against the Delta Variant: A Retrospective Cohort Study, Clinical Infectious Diseases,
doi:10.1093/CID/CIAB999Kodaka, A new cell-based assay to evaluate myogenesis in mouse myoblast C2C12 cells, Experimental cell research
Lan, Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor, Nature
Liu, Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization, Cell host & microbe
Lopez Bernal, Effectiveness of Covid-19 vaccines against the B. 1.617. 2 (Delta) variant, N Engl J Med
Mccallum, N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2, Cell
Mccarthy, Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape, Science
Meng, Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B. 1.1. 7, Cell reports
Meng, SARS-CoV-2 Omicron spike mediated immune escape, infectivity and cell-cell fusion
Mykytyn, SARS-CoV-2 entry into human airway organoids is serine proteasemediated and facilitated by the multibasic cleavage site, Elife
Newman, Neutralising antibody activity against SARS-CoV-2 variants, including Omicron, in an elderly cohort vaccinated with BNT162b2, medRxiv,
doi:10.1101/2021.12.23.21268293Nie, Functional comparison of SARS-CoV-2 with closely related pangolin and bat coronaviruses, Cell discovery
Papa, Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion, PLoS pathogens
Peacock, The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets, Nature Microbiology
Pinto, Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody, Nature
Pymol, The PyMOL molecular graphics system, Version
Rihn, A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research, PLoS biology
Rouet, Potent SARS-CoV-2 binding and neutralization through maturation of iconic SARS-CoV-1 antibodies, Mabs
Saccon, Cell-type-resolved quantitative proteomics map of interferon response against SARS-CoV-2, Iscience
Sheikh, Kerr, Woolhouse, Mcmenamin, Robertson, against symptomatic disease: national cohort with nested test negative design study in Scotland
Starr, Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding, Cell
Starr, Prospective mapping of viral mutations that escape antibodies used to treat COVID-19, Science
Sweredoski, Baldi, PEPITO: improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure, Bioinformatics
Thorne, Evolution of enhanced innate immune evasion by SARS-CoV-2, Nature
Viana, Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa
Voysey, Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK, The Lancet
Wall, AZD1222-induced neutralising antibody activity against SARS-CoV-2 Delta VOC, The Lancet
Wall, Neutralising antibody activity against SARS-CoV-2 VOCs B. 1.617. 2 and B. 1.351 by BNT162b2 vaccination, The Lancet
Wang, mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants, Nature
Weisblum, Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants, Elife
Winstone, The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2, Journal of virology
Woo, Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane, The Journal of Physical Chemistry B
Wood, Generalized additive models: an introduction with R
Wrapp, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science
Wrobel, Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2, Nature communications
Zahradník, SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution, Nature microbiology
Zhang, Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant, Science
Zufferey, Nagy, Mandel, Naldini, Trono, Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo, Nature biotechnology
Zufferey, Self-Inactivating Lentivirus Vector for Safe and Efficient In Vivo Gene Delivery, Journal of Virology
DOI record:
{
"DOI": "10.1101/2022.01.03.21268111",
"URL": "http://dx.doi.org/10.1101/2022.01.03.21268111",
"abstract": "<jats:p>Vaccination-based exposure to spike protein derived from early SARS-CoV-2 sequences is the key public health strategy against COVID-19. Successive waves of SARS-CoV-2 infections have been characterised by the evolution of highly mutated variants that are more transmissible and that partially evade the adaptive immune response. Omicron is the fifth of these Variants of Concern (VOCs) and is characterised by a step change in transmission capability, suggesting significant antigenic and biological change. It is characterised by 45 amino acid substitutions, including 30 changes in the spike protein relative to one of the earliest sequences, Wuhan-Hu-1, of which 15 occur in the receptor-binding domain, an area strongly associated with humoral immune evasion. In this study, we demonstrate both markedly decreased neutralisation in serology assays and real-world vaccine effectiveness in recipients of two doses of vaccine, with efficacy partially recovered by a third mRNA booster dose. We also show that immunity from natural infection (without vaccination) is more protective than two doses of vaccine but inferior to three doses. Finally, we demonstrate fundamental changes in the Omicron entry process in vitro, towards TMPRSS2-independent fusion, representing a major shift in the replication properties of SARS-CoV-2. Overall, these findings underlie rapid global transmission and may alter the clinical severity of disease associated with the Omicron variant.</jats:p>",
"accepted": {
"date-parts": [
[
2022,
1,
3
]
]
},
"author": [
{
"affiliation": [],
"family": "Willett",
"given": "Brian J",
"sequence": "first"
},
{
"affiliation": [],
"family": "Grove",
"given": "Joe",
"sequence": "additional"
},
{
"affiliation": [],
"family": "MacLean",
"given": "Oscar",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wilkie",
"given": "Craig",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Logan",
"given": "Nicola",
"sequence": "additional"
},
{
"affiliation": [],
"family": "De Lorenzo",
"given": "Giuditta",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Furnon",
"given": "Wilhelm",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Scott",
"given": "Sam",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Manali",
"given": "Maria",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Szemiel",
"given": "Agnieszka",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ashraf",
"given": "Shirin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Vink",
"given": "Elen",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Harvey",
"given": "William",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Davis",
"given": "Chris",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Orton",
"given": "Richard",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hughes",
"given": "Joseph",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Holland",
"given": "Poppy",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Silva",
"given": "Sofia Vanessa",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pascall",
"given": "David",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Puxty",
"given": "Kathryn",
"sequence": "additional"
},
{
"affiliation": [],
"family": "da Silva Filipe",
"given": "Ana",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yebra",
"given": "Gonzalo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Shaaban",
"given": "Sharif",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Holden",
"given": "Matthew T.G.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pinto",
"given": "Rute Maria",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Gunson",
"given": "Rory",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Templeton",
"given": "Kate",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Murcia",
"given": "Pablo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Patel",
"given": "Arvind",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Haughney",
"given": "John",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Robertson",
"given": "David L",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Palmarini",
"given": "Massimo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ray",
"given": "Surajit",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1482-0889",
"affiliation": [],
"authenticated-orcid": false,
"family": "Thomson",
"given": "Emma C",
"sequence": "additional"
},
{
"affiliation": [],
"name": "The COVID-19 DeplOyed VaccinE (DOVE) Investigators",
"sequence": "additional"
},
{
"affiliation": [],
"name": "COVID-19 EVADE Cohort Study",
"sequence": "additional"
},
{
"affiliation": [],
"name": "The COVID-19 Genomics UK (COG-UK) Consortium",
"sequence": "additional"
},
{
"affiliation": [],
"name": "The G2P-UK National Virology Consortium",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
1,
4
]
],
"date-time": "2022-01-04T06:40:15Z",
"timestamp": 1641278415000
},
"deposited": {
"date-parts": [
[
2022,
1,
4
]
],
"date-time": "2022-01-04T06:40:16Z",
"timestamp": 1641278416000
},
"group-title": "Infectious Diseases (except HIV/AIDS)",
"indexed": {
"date-parts": [
[
2022,
1,
4
]
],
"date-time": "2022-01-04T07:15:29Z",
"timestamp": 1641280529641
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2022,
1,
3
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2022.01.03.21268111",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2022,
1,
3
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2022,
1,
3
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference-count": 0,
"references-count": 0,
"relation": {},
"score": 1,
"short-container-title": [],
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": [
"The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism"
],
"type": "posted-content"
}