Highly Specific Sigma Receptor Ligands Exhibit Anti-Viral Properties in SARS-CoV-2 Infected Cells
David A Ostrov, Andrew P Bluhm, Danmeng Li, Juveriya Qamar Khan, Megha Rohamare, Karthic Rajamanickam, Kalpana K. Bhanumathy, Jocelyne Lew, Darryl Falzarano, Franco J Vizeacoumar, Joyce A Wilson, Marco Mottinelli, Siva Rama Raju Kanumuri, Abhisheak Sharma, Christopher R Mccurdy, Michael H Norris
Pathogens, doi:10.3390/pathogens10111514
1) Background: There is a strong need for prevention and treatment strategies for COVID-19 that are not impacted by SARS-CoV-2 mutations emerging in variants of concern. After virus infection, host ER resident sigma receptors form direct interactions with non-structural SARS-CoV-2 proteins present in the replication complex. (2) Methods: In this work, highly specific sigma receptor ligands were investigated for their ability to inhibit both SARS-CoV-2 genome replication and virus induced cellular toxicity. This study found antiviral activity associated with agonism of the sigma-1 receptor (e.g., SA4503), ligation of the sigma-2 receptor (e.g., CM398), and a combination of the two pathways (e.g., AZ66). ( 3 ) Results: Intermolecular contacts between these ligands and sigma receptors were identified by structural modeling. (4) Conclusions: Sigma receptor ligands and drugs with off-target sigma receptor binding characteristics were effective at inhibiting SARS-CoV-2 infection in primate and human cells, representing a potential therapeutic avenue for COVID-19 prevention and treatment.
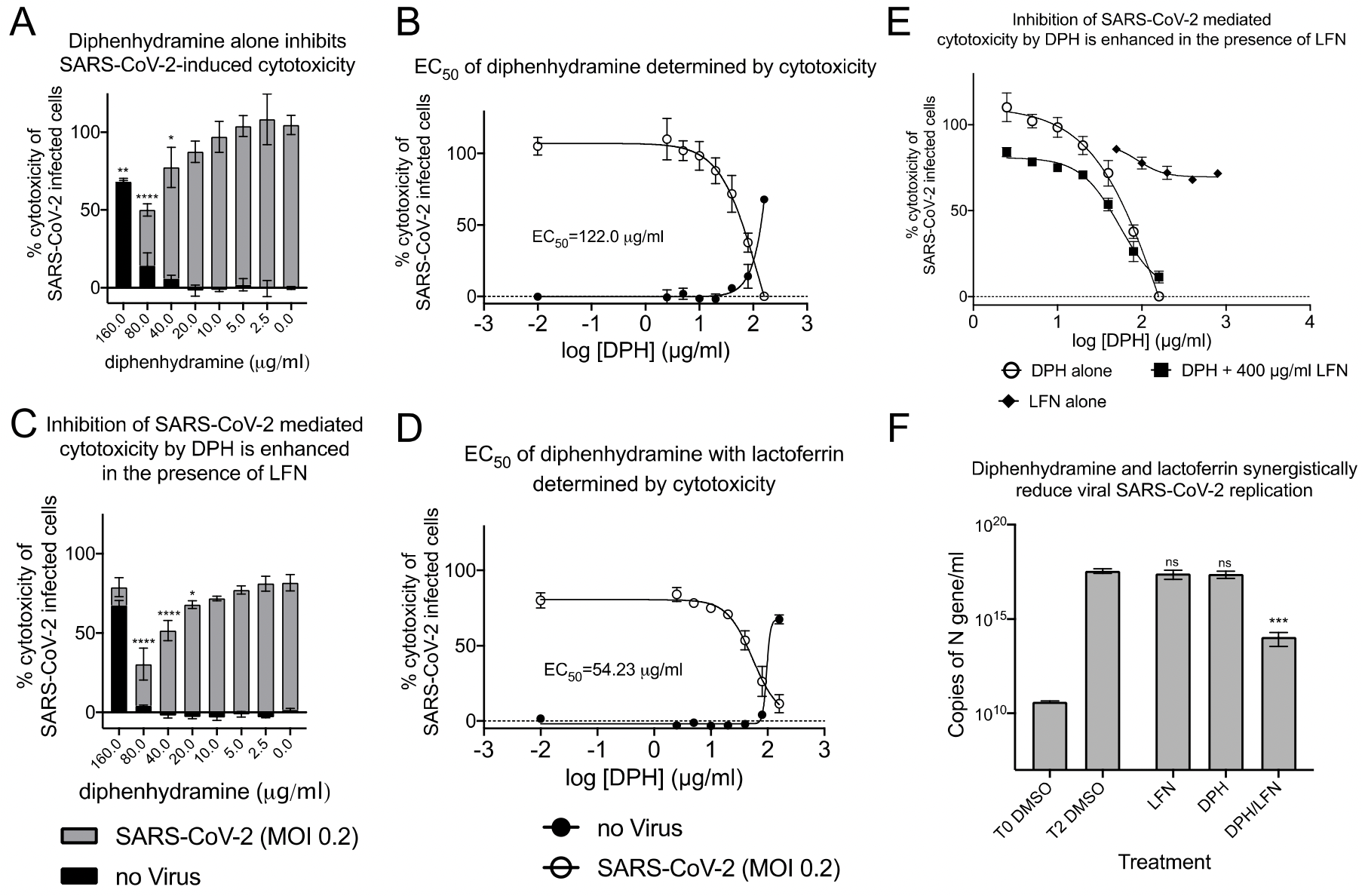
Controls included total LDH release as measured by lysis of all cells, spontaneous release from uninfected cells, and media alone. The toxicity of sigma ligands alone were also determined in parallel to discriminate the amount of SARS-CoV-2-induced cytotoxicity occurring in the presence of a given treatment. After spontaneous and background subtraction, OD 450 values were transformed to a percent of SARS-CoV-2 infected cells (100%) in the absence of any drug treatment to obtain percent of SARS-CoV-2-induced cytotoxicity. These experiments were carried out twice.
Plaque Reduction Assay Vero E6 cells were plated in 24-well plates with triplicate replicates on different plates. Virus master mix was used to dilute down to 20-200 PFU/mL and aliquoted in separate tubes with drugs at the final indicated drug concentrations. The drug virus mixtures were immediately used to infect Vero E6 monolayers for 1 h with rocking every 10 min. Monolayers were then overlaid with MEM in 1.5% low-melt agarose containing drugs at the final concentrations indicated. Plaques were counted at 72 hours post infection and used to calculate the apparent reduction in viral concentration compared to the starting volume. Data presented is representative of two independent experiments.
Generation of ACE-2 Lentivirus Particles The lentivirus containing ACE2 were generated by co-transfecting psPAX2, pMD2.G, and an ACE expression vector that also contained a blasticidin selection gene EX-U1285-Lv197..
References
Abate, Niso, Berardi, Sigma-2 receptor: Past, present and perspectives on multiple therapeutic exploitations, Futur. Med. Chem,
doi:10.4155/fmc-2018-0072Arnold, Bordoli, Kopp, Schwede, The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling, Bioinformatics,
doi:10.1093/bioinformatics/bti770Basile, Paul, Mirchevich, Kuijpers, De Costa, Modulation of (+)-[3H]pentazocine binding to guinea pig cerebellum by divalent cations, Mol. Pharmacol
Cirino, Eans, Medina, Wilson, Mottinelli et al., Characterization of Sigma 1 Receptor Antagonist CM-304 and Its Analog, AZ-66: Novel Therapeutics Against Allodynia and Induced Pain, Front. Pharmacol,
doi:10.3389/fphar.2019.00678Cunningham, Li, Vizeacoumar, Bhanumathy, Lee et al., Therapeutic relevance of the protein phosphatase 2A in cancer, Oncotarget,
doi:10.18632/oncotarget.11399Fung, Huang, Liu, Coronavirus-induced ER stress response and its involvement in regulation of coronavirus-host interactions, Virus Res,
doi:10.1016/j.virusres.2014.09.016Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature,
doi:10.1038/s41586-020-2286-9Hernández, Gabrielli, Costa, Uttaro, Phagocytic and pinocytic uptake of cholesterol in Tetrahymena thermophila impact differently on gene regulation for sterol homeostasis, Sci. Rep,
doi:10.1038/s41598-021-88737-zHu, Meng, Zhang, Xiang, Wang, The in vitro antiviral activity of lactoferrin against common human coronaviruses and SARS-CoV-2 is mediated by targeting the heparan sulfate co-receptor, Emerg. Microbes Infect,
doi:10.1080/22221751.2021.1888660Intagliata, Sharma, King, Mesangeau, Seminerio et al., Discovery of a Highly Selective Sigma-2 Receptor Ligand, 1-(4-(6,7-Dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)butyl)-3-methyl-1H-benzo[d]imidazol-2(3H)-one (CM398), with Drug-Like Properties and Antinociceptive Effects In Vivo, AAPS J,
doi:10.1208/s12248-020-00472-xJamalapuram, Vuppala, Abdelazeem, Mccurdy, Avery, Ultra-performance liquid chromatography tandem mass spectrometry method for the determination of AZ66, a sigma receptor ligand, in rat plasma and its application to in vivo pharmacokinetics, Biomed. Chromatogr,
doi:10.1002/bmc.2901James, Shen, Zavaleta, Nielsen, Mesangeau et al., New Positron Emission Tomography (PET) Radioligand for Imaging σ-1 Receptors in Living Subjects, J. Med. Chem,
doi:10.1021/jm300371cJang, Jeon, Kim, Lee, Drugs repurposed for COVID-19 by virtual screening of 6218 drugs and cell-based assay, Proc. Natl. Acad. Sci,
doi:10.1073/pnas.2024302118Jeon, Ko, Lee, Choi, Byun et al., Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs, Antimicrob. Agents Chemother,
doi:10.1128/AAC.00819-20Joshi, Joshi, Degani, Tackling SARS-CoV-2: Proposed targets and repurposed drugs, Futur. Med. Chem,
doi:10.4155/fmc-2020-0147Kawamura, Ishiwata, Tajima, Ishii, Matsuno et al., In vivo evaluation of [11C]SA4503 as a PET ligand for mapping CNS sigma1 receptors, Nucl. Med. Biol,
doi:10.1016/S0969-8051(00)00081-0Lane, Ekins, Defending Antiviral Cationic Amphiphilic Drugs That May Cause Drug-Induced Phospholipidosis, J. Chem. Inf. Model,
doi:10.1021/acs.jcim.1c00903Li, Han, Dai, Xu, He et al., Distinct mechanisms for TMPRSS2 expression explain organ-specific inhibition of SARS-CoV-2 infection by enzalutamide, Nat. Commun,
doi:10.1038/s41467-021-21171-xLi, Zhang, Yao, Xiang, Ma et al., Sigma-1 receptor agonist increases axon outgrowth of hippocampal neurons via voltage-gated calcium ions channels, CNS Neurosci. Ther,
doi:10.1111/cns.12768Long, Hassan, Thompson, Mcdonald, Wang et al., Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition, Nat. Commun,
doi:10.1038/s41467-019-10279-wMatsuno, Nakazawa, Okamoto, Kawashima, Mita, Binding properties of SA4503, a novel and selective sigma 1 receptor agonist, Eur J. Pharmacol,
doi:10.1016/0014-2999(96)00201-4Mirabelli, Wotring, Zhang, Mccarty, Fursmidt et al., Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19, bioRxiv,
doi:10.1073/pnas.2105815118Morris, Huey, Lindstrom, Sanner, Belew et al., AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem,
doi:10.1002/jcc.21256Mésangeau, Narayanan, Green, Shaikh, Kaushal et al., Conversion of a Highly Selective Sigma-1 Receptor-Ligand to Sigma-2 Receptor Preferring Ligands with Anticocaine Activity, J. Med. Chem,
doi:10.1021/jm701357mPoduri, Joshi, Jagadeesh, Drugs targeting various stages of the SARS-CoV-2 life cycle: Exploring promising drugs for the treatment of covid-19, Cell Signal,
doi:10.1016/j.cellsig.2020.109721Reznikov, Norris, Vashisht, Bluhm, Li et al., Identification of antiviral antihistamines for COVID-19 repurposing, Biochem. Biophys. Res. Commun,
doi:10.1016/j.bbrc.2020.11.095Riva, Yuan, Yin, Martin-Sancho, Matsunaga et al., Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing, Nature
Seminerio, Robson, Abdelazeem, Mesangeau, Jamalapuram et al., Synthesis and Pharmacological Characterization of a Novel Sigma Receptor Ligand with Improved Metabolic Stability and Antagonistic Effects Against Methamphetamine, AAPS J,
doi:10.1208/s12248-011-9311-8Shi, Shuai, Wen, Wang, Yan et al., The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract, Emerg. Microbes Infect,
doi:10.1080/22221751.2021.1899770Tummino, Rezelj, Fischer, Fischer, O'meara et al., Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2, Science,
doi:10.1126/science.abi4708Xu, Kaushal, Shaikh, Wilson, Mésangeau et al., A Novel Substituted Piperazine, CM156, Attenuates the Stimulant and Toxic Effects of Cocaine in Mice, J. Pharmacol. Exp. Ther,
doi:10.1124/jpet.109.161398Yang, Zeng, Wang, Sun, Yin et al., Sigma-2 Receptor-A Potential Target for Cancer/Alzheimer's Disease Treatment via Its Regulation of Cholesterol Homeostasis, Molecules,
doi:10.3390/molecules25225439Zeng, Riad, Mach, The Biological Function of Sigma-2 Receptor/TMEM97 and Its Utility in PET Imaging Studies in Cancer, Cancers,
doi:10.3390/cancers12071877Zheng, Ma, Xiong, Fan, Efficacy and safety of direct acting antiviral regimens for hepatitis C virus and human immunodeficiency virus co-infection: Systematic review and network meta-analysis, J. Gastroenterol. Hepatol
Zhou, Hou, Shen, Huang, Martin et al., Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2, Cell Discov,
doi:10.1038/s41421-020-0153-3DOI record:
{
"DOI": "10.3390/pathogens10111514",
"ISSN": [
"2076-0817"
],
"URL": "http://dx.doi.org/10.3390/pathogens10111514",
"abstract": "<jats:p>(1) Background: There is a strong need for prevention and treatment strategies for COVID-19 that are not impacted by SARS-CoV-2 mutations emerging in variants of concern. After virus infection, host ER resident sigma receptors form direct interactions with non-structural SARS-CoV-2 proteins present in the replication complex. (2) Methods: In this work, highly specific sigma receptor ligands were investigated for their ability to inhibit both SARS-CoV-2 genome replication and virus induced cellular toxicity. This study found antiviral activity associated with agonism of the sigma-1 receptor (e.g., SA4503), ligation of the sigma-2 receptor (e.g., CM398), and a combination of the two pathways (e.g., AZ66). (3) Results: Intermolecular contacts between these ligands and sigma receptors were identified by structural modeling. (4) Conclusions: Sigma receptor ligands and drugs with off-target sigma receptor binding characteristics were effective at inhibiting SARS-CoV-2 infection in primate and human cells, representing a potential therapeutic avenue for COVID-19 prevention and treatment.</jats:p>",
"alternative-id": [
"pathogens10111514"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-4696-875X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ostrov",
"given": "David A.",
"sequence": "first"
},
{
"affiliation": [],
"family": "Bluhm",
"given": "Andrew P.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6905-554X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Li",
"given": "Danmeng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Khan",
"given": "Juveriya Qamar",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rohamare",
"given": "Megha",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rajamanickam",
"given": "Karthic",
"sequence": "additional"
},
{
"affiliation": [],
"family": "K. Bhanumathy",
"given": "Kalpana",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lew",
"given": "Jocelyne",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8805-8068",
"affiliation": [],
"authenticated-orcid": false,
"family": "Falzarano",
"given": "Darryl",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Vizeacoumar",
"given": "Franco J.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5051-3088",
"affiliation": [],
"authenticated-orcid": false,
"family": "Wilson",
"given": "Joyce A.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5725-0439",
"affiliation": [],
"authenticated-orcid": false,
"family": "Mottinelli",
"given": "Marco",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6087-6587",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kanumuri",
"given": "Siva Rama Raju",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0553-4039",
"affiliation": [],
"authenticated-orcid": false,
"family": "Sharma",
"given": "Abhisheak",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8695-2915",
"affiliation": [],
"authenticated-orcid": false,
"family": "McCurdy",
"given": "Christopher R.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5285-0432",
"affiliation": [],
"authenticated-orcid": false,
"family": "Norris",
"given": "Michael H.",
"sequence": "additional"
}
],
"container-title": [
"Pathogens"
],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
11,
22
]
],
"date-time": "2021-11-22T02:00:50Z",
"timestamp": 1637546450000
},
"deposited": {
"date-parts": [
[
2021,
12,
1
]
],
"date-time": "2021-12-01T05:06:20Z",
"timestamp": 1638335180000
},
"indexed": {
"date-parts": [
[
2021,
12,
11
]
],
"date-time": "2021-12-11T19:44:47Z",
"timestamp": 1639251887709
},
"is-referenced-by-count": 0,
"issn-type": [
{
"type": "electronic",
"value": "2076-0817"
}
],
"issue": "11",
"issued": {
"date-parts": [
[
2021,
11,
20
]
]
},
"journal-issue": {
"issue": "11",
"published-online": {
"date-parts": [
[
2021,
11
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
11,
20
]
],
"date-time": "2021-11-20T00:00:00Z",
"timestamp": 1637366400000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2076-0817/10/11/1514/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1514",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2021,
11,
20
]
]
},
"published-online": {
"date-parts": [
[
2021,
11,
20
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1128/AAC.00819-20",
"doi-asserted-by": "publisher",
"key": "ref1"
},
{
"DOI": "10.4155/fmc-2020-0147",
"doi-asserted-by": "publisher",
"key": "ref2"
},
{
"DOI": "10.1038/s41421-020-0153-3",
"doi-asserted-by": "publisher",
"key": "ref3"
},
{
"DOI": "10.1016/j.cellsig.2020.109721",
"doi-asserted-by": "publisher",
"key": "ref4"
},
{
"DOI": "10.1038/s41586-020-2577-1",
"doi-asserted-by": "publisher",
"key": "ref5"
},
{
"DOI": "10.1073/pnas.2024302118",
"doi-asserted-by": "publisher",
"key": "ref6"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"doi-asserted-by": "publisher",
"key": "ref7"
},
{
"DOI": "10.1016/j.bbrc.2020.11.095",
"doi-asserted-by": "publisher",
"key": "ref8"
},
{
"DOI": "10.1021/acs.jmedchem.0c00502",
"doi-asserted-by": "publisher",
"key": "ref9"
},
{
"DOI": "10.3389/fphar.2020.582310",
"doi-asserted-by": "publisher",
"key": "ref10"
},
{
"DOI": "10.4155/fmc-2018-0072",
"doi-asserted-by": "publisher",
"key": "ref11"
},
{
"DOI": "10.3390/cancers12071877",
"doi-asserted-by": "publisher",
"key": "ref12"
},
{
"DOI": "10.3390/molecules25225439",
"doi-asserted-by": "publisher",
"key": "ref13"
},
{
"DOI": "10.1111/cns.12768",
"doi-asserted-by": "publisher",
"key": "ref14"
},
{
"DOI": "10.1016/S0969-8051(00)00081-0",
"doi-asserted-by": "publisher",
"key": "ref15"
},
{
"DOI": "10.1016/0014-2999(96)00201-4",
"doi-asserted-by": "publisher",
"key": "ref16"
},
{
"DOI": "10.3389/fphar.2019.00678",
"doi-asserted-by": "publisher",
"key": "ref17"
},
{
"DOI": "10.1208/s12248-020-00472-x",
"doi-asserted-by": "publisher",
"key": "ref18"
},
{
"DOI": "10.1002/bmc.2901",
"doi-asserted-by": "publisher",
"key": "ref19"
},
{
"DOI": "10.1124/jpet.109.161398",
"doi-asserted-by": "publisher",
"key": "ref20"
},
{
"DOI": "10.1021/jm701357m",
"doi-asserted-by": "publisher",
"key": "ref21"
},
{
"DOI": "10.1208/s12248-011-9311-8",
"doi-asserted-by": "publisher",
"key": "ref22"
},
{
"DOI": "10.1080/22221751.2021.1899770",
"doi-asserted-by": "publisher",
"key": "ref23"
},
{
"DOI": "10.1093/bioinformatics/bti770",
"doi-asserted-by": "publisher",
"key": "ref24"
},
{
"DOI": "10.1038/s41467-019-10279-w",
"doi-asserted-by": "publisher",
"key": "ref25"
},
{
"DOI": "10.1038/s41598-021-88737-z",
"doi-asserted-by": "publisher",
"key": "ref26"
},
{
"article-title": "Modulation of (+)-[3H]pentazocine binding to guinea pig cerebellum by divalent cations",
"author": "Basile",
"first-page": "882",
"journal-title": "Mol. Pharmacol.",
"key": "ref27",
"volume": "42",
"year": "1992"
},
{
"DOI": "10.1073/pnas.2105815118",
"doi-asserted-by": "publisher",
"key": "ref28"
},
{
"DOI": "10.1080/22221751.2021.1888660",
"doi-asserted-by": "publisher",
"key": "ref29"
},
{
"DOI": "10.1038/s41467-021-21171-x",
"doi-asserted-by": "publisher",
"key": "ref30"
},
{
"DOI": "10.1146/annurev-virology-100114-055218",
"doi-asserted-by": "publisher",
"key": "ref31"
},
{
"DOI": "10.3389/fmicb.2014.00296",
"doi-asserted-by": "publisher",
"key": "ref32"
},
{
"DOI": "10.1016/j.virusres.2014.09.016",
"doi-asserted-by": "publisher",
"key": "ref33"
},
{
"DOI": "10.3389/fnins.2019.00733",
"doi-asserted-by": "publisher",
"key": "ref34"
},
{
"DOI": "10.1126/science.abi4708",
"doi-asserted-by": "publisher",
"key": "ref35"
},
{
"DOI": "10.1111/jgh.15051",
"doi-asserted-by": "publisher",
"key": "ref36"
},
{
"DOI": "10.1177/1087057111413919",
"doi-asserted-by": "publisher",
"key": "ref37"
},
{
"DOI": "10.1021/acs.jcim.1c00903",
"doi-asserted-by": "publisher",
"key": "ref38"
},
{
"DOI": "10.1021/jm300371c",
"doi-asserted-by": "publisher",
"key": "ref39"
},
{
"DOI": "10.18632/oncotarget.11399",
"doi-asserted-by": "publisher",
"key": "ref40"
},
{
"DOI": "10.1002/jcc.21256",
"doi-asserted-by": "publisher",
"key": "ref41"
}
],
"reference-count": 41,
"references-count": 41,
"relation": {},
"score": 1,
"short-container-title": [
"Pathogens"
],
"short-title": [],
"source": "Crossref",
"subject": [
"Infectious Diseases",
"Microbiology (medical)",
"General Immunology and Microbiology",
"Molecular Biology",
"Immunology and Allergy"
],
"subtitle": [],
"title": [
"Highly Specific Sigma Receptor Ligands Exhibit Anti-Viral Properties in SARS-CoV-2 Infected Cells"
],
"type": "journal-article",
"volume": "10"
}