SARS-CoV-2 Genetic Variants and Patient Factors Associated with Hospitalization Risk
Tonia Korves, David Stein, David Walburger, Tomasz Adamusiak, Seth Roberts
doi:10.1101/2024.03.08.24303818
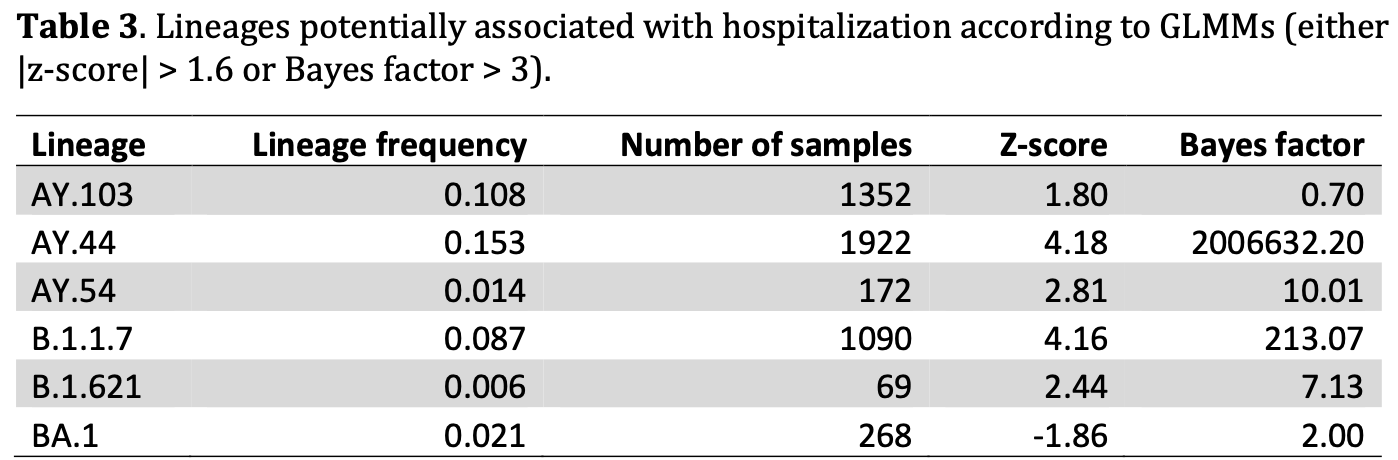
Variants of SARS-CoV-2 have been associated with different transmissibilities and disease severities. The present study examines SARS-CoV-2 genetic variants and their relationship to risk for hospitalization, using data from 12,538 patients from a large, multisite observational cohort study. The association of viral genomic variants and hospitalization is examined with clinical covariates, including COVID-19 vaccination status, outpatient monoclonal antibody treatment status, and underlying risk for poor clinical outcome. Modeling approaches include XGBoost with SHapley Additive exPlanations (SHAP) analysis and generalized linear mixed models. The results indicate that several SARS-CoV-2 lineages are associated with increased hospitalization risk, including B.1.1.7, AY.44, and AY.54. As found in prior studies, Omicron is associated with lower hospitalization risk compared to prior WHO variants. In addition, the results suggest that variants at specific amino acid locations, including locations within Spike protein N-terminal domain and in non-structural protein 14, are associated with hospitalization risk.
Competing Interests There are no competing interests to declare.
References
Agarwal, Leblond, Mcauley, Linking genotype to phenotype: Further exploration of mutations in SARS-CoV-2 associated with mild or severe outcomes, Internet,
doi:10.1101/2022.04.15.22273922v1Aiewsakun, Nilplub, Wongtrakoongate, SARS-CoV-2 genetic variations associated with COVID-19 pathogenicity [Internet], Microbial Genomics,
doi:10.1099/mgen.0.000734Aksamentov, Roemer, Hodcroft, Nextclade: Clade assignment, mutation calling and quality control for viral genomes [Internet, Journal of Open Source Software,
doi:10.21105/joss.03773Ambrose, Amin, Anderson, Neutralizing monoclonal antibody use and COVID-19 infection outcomes [Internet, JAMA Network Open
Berman, Westbrook, Feng, The protein data bank, Nucleic Acids Research
Bingham, Chen, Jankowiak, Pyro: Deep universal probabilistic programming [Internet, Journal of Machine Learning Research
Carabelli, Peacock, Thorne, SARS-CoV-2 variant biology: Immune escape, transmission and fitness [Internet], Nature reviews Microbiology
Chen, Guestrin, XGBoost: A scalable tree boosting system
Cingolani, Platts, Wang, A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso-3 [Internet, Fly,
doi:10.4161/fly.19695Elixhauser, Steiner, Harris, Comorbidity measures for use with administrative data [Internet], Medical care
Esper, Cheng, Adhikari, Genomic epidemiology of SARS-CoV-2 infection during the initial pandemic wave and association with disease severity [Internet, JAMA network open
Goyal, Bruyne, Belkum A Van, Different SARS-CoV-2 haplotypes associate with geographic origin and case fatality rates of COVID-19 patients, Infection, Genetics and Evolution
Grubaugh, Gangavarapu, Quick, An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar, bioRxiv,
doi:10.1101/383513v1Hahn, Wu, Lee, Genome-wide association analysis of COVID-19 mortality risk in SARS-CoV-2 genomes identifies mutation in the SARS-CoV-2 spike protein that colocalizes with p.1 of the Brazilian strain [Internet], Genetic Epidemiology,
doi:10.1002/gepi.22421Hsu, Laurent-Rolle, Pawlak, Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein [Internet, Proceedings of the National Academy of Sciences of the United States of America,
doi:10.1073/pnas.2101161118Johnsen, Riemer-Sørensen, Dewan, A new method for exploring genegene and gene-environment interactions in GWAS with tree ensemble methods and SHAP © 2024 The MITRE Corporation, All Rights Reserved Approved for Public Release. Distribution Unlimited,
doi:10.1186/s12859-021-04041-7Johnsen, Strümke, Langaas, Inferring feature importance with uncertainties with application to large genotype data [Internet], PLOS Computational Biology,
doi:10.1371/journal.pcbi.1010963Jumper, Evans, Pritzel, Highly accurate protein structure prediction with AlphaFold [Internet], Nature
Khare, Gurry, Freitas, GISAID's role in pandemic response [Internet], China CDC Weekly
Kind, Buckingham, Making neighborhood-disadvantage metrics accessible -the neighborhood atlas, New England Journal of Medicine
Koch, Du, Dressner, Demographic and viral-genetic analyses of COVID-19 severity in Bahrain identify local risk factors and a protective effect of polymerase mutations
Krivov, Shapovalov, Dunbrack, Improved prediction of protein side-chain conformations with SCWRL4 [Internet], Proteins: Structure, Function, and Bioinformatics,
doi:10.1002/prot.22488Liang, Ding, Liu, Identification of critical SARS-CoV-2 amino acids associated with COVID-19 hospitalization rate using machine learning and statistical modeling: An observational study in the United States [Internet]. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases
Lundberg, Allen, Lee, A unified approach to interpreting model predictions, Advances in Neural Information Processing Systems
Maher, Bartha, Weaver, Predicting the mutational drivers of future SARS-CoV-2 variants of concern [Internet], Science translational medicine
Maurya, Mishra, Swaminathan, SARS-CoV-2 mutations and COVID-19 clinical outcome: Mutation global frequency dynamics and structural modulation hold the key [Internet], Frontiers in cellular and infection microbiology
Mayer, SHAP visualizations [r package shapviz version 0
Mehta, Alle, Chaturvedi, Clinico-genomic analysis reveals mutations associated with COVID-19 disease severity: Possible modulation by RNA structure [Internet, Pathogens
Meng, Goddard, Pettersen, UCSF ChimeraX: Tools for structure building and analysis [Internet], Protein Science,
doi:10.1002/pro.4792Nagy, Pongor, Győrffy, Different mutations in SARS-CoV-2 associate with severe and mild outcome [Internet, International journal of antimicrobial agents
O'toole, Scher, Underwood, Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool [Internet, Virus Evolution
Obermeyer, Jankowiak, Barkas, Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness [Internet], Science,
doi:10.1126/science.abm1208Ramarao-Milne, Jain, Sng, Data-driven platform for identifying variants of interest in COVID-19 virus [Internet], Computational and structural biotechnology journal
Rouder, Speckman, Sun, Bayesian t tests for accepting and rejecting the null hypothesis [Internet], Psychonomic Bulletin and Review,
doi:10.3758/PBR.16.2.225Shen, Triche, Bard, Spike protein NTD mutation G142D in SARS-CoV-2 delta VOC lineages is associated with frequent back mutations, increased viral loads, and immune evasion, Internet,
doi:10.1101/2021.09.12.21263475v1Sokhansanj, Zhao, Rosen, An interpretable deep learning model for predicting the risk of severe COVID-19 from spike protein sequence
Turakhia, Thornlow, Hinrichs, Ultrafast sample placement on existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic [Internet], Nature Genetics
Verdinelli, Wasserman, Computing bayes factors using a generalization of the savage-dickey density ratio, Journal of the American Statistical Association
Voss, Skarzynski, Mcauley, Variants in SARS-CoV-2 associated with mild or severe outcome [Internet], Evolution, Medicine, and Public Health
Wang, Chen, Hozumi, Decoding asymptomatic COVID-19 infection and transmission [Internet], Journal of Physical Chemistry Letters,
doi:10.1021/acs.jpclett.0c02765Wingate, Weber, Automated variational inference in probabilistic programming
Wu, Zhao, Yu, A new coronavirus associated with human respiratory disease in china [Internet], Nature
Yan, Machine learning evaluation metrics, r package MLmetrics version
Zhang, Tan, Ling, Viral and host factors related to the clinical outcome of COVID-19 [Internet], Nature
Zhu, Marsh, Griffith, Predictive model for severe COVID-19 using SARS-CoV-2 whole-genome sequencing and electronic health record data, March 2020-may 2021, PloS one
DOI record:
{
"DOI": "10.1101/2024.03.08.24303818",
"URL": "http://dx.doi.org/10.1101/2024.03.08.24303818",
"abstract": "<jats:title>ABSTRACT</jats:title><jats:p>Variants of SARS-CoV-2 have been associated with different transmissibilities and disease severities. The present study examines SARS-CoV-2 genetic variants and their relationship to risk for hospitalization, using data from 12,538 patients from a large, multisite observational cohort study. The association of viral genomic variants and hospitalization is examined with clinical covariates, including COVID-19 vaccination status, outpatient monoclonal antibody treatment status, and underlying risk for poor clinical outcome. Modeling approaches include XGBoost with SHapley Additive exPlanations (SHAP) analysis and generalized linear mixed models. The results indicate that several SARS-CoV-2 lineages are associated with increased hospitalization risk, including B.1.1.7, AY.44, and AY.54. As found in prior studies, Omicron is associated with lower hospitalization risk compared to prior WHO variants. In addition, the results suggest that variants at specific amino acid locations, including locations within Spike protein N-terminal domain and in non-structural protein 14, are associated with hospitalization risk.</jats:p>",
"accepted": {
"date-parts": [
[
2024,
3,
10
]
]
},
"author": [
{
"affiliation": [],
"family": "Korves",
"given": "Tonia",
"sequence": "first"
},
{
"affiliation": [],
"family": "Stein",
"given": "David",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Walburger",
"given": "David",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Adamusiak",
"given": "Tomasz",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Roberts",
"given": "Seth",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
3,
11
]
],
"date-time": "2024-03-11T05:20:14Z",
"timestamp": 1710134414000
},
"deposited": {
"date-parts": [
[
2024,
3,
13
]
],
"date-time": "2024-03-13T07:41:29Z",
"timestamp": 1710315689000
},
"group-title": "Infectious Diseases (except HIV/AIDS)",
"indexed": {
"date-parts": [
[
2024,
3,
14
]
],
"date-time": "2024-03-14T00:49:02Z",
"timestamp": 1710377342975
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
3,
10
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2024.03.08.24303818",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2024,
3,
10
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2024,
3,
10
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"article-title": "SARS-CoV-2 variant biology: Immune escape, transmission and fitness [Internet]",
"first-page": "162",
"journal-title": "Nature reviews Microbiology",
"key": "2024031300400911000_2024.03.08.24303818v1.1",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1016/j.ijantimicag.2020.106272",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.2",
"unstructured": "Nagy Á , Pongor S , Győrffy B: Different mutations in SARS-CoV-2 associate with severe and mild outcome [Internet]. International journal of antimicrobial agents 2021; 57 Available from: https://pubmed.ncbi.nlm.nih.gov/33347989/"
},
{
"DOI": "10.1001/jamanetworkopen.2021.7746",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.3",
"unstructured": "Esper FP , Cheng YW , Adhikari TM , et al.: Genomic epidemiology of SARS-CoV-2 infection during the initial pandemic wave and association with disease severity [Internet]. JAMA network open 2021; 4 Available from: https://pubmed.ncbi.nlm.nih.gov/33900399/"
},
{
"DOI": "10.1002/gepi.22421",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.4"
},
{
"DOI": "10.1093/emph/eoab019",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.5"
},
{
"DOI": "10.1099/mgen.0.000734",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.6",
"unstructured": "Aiewsakun P , Nilplub P , Wongtrakoongate P , et al.: SARS-CoV-2 genetic variations associated with COVID-19 pathogenicity [Internet]. Microbial Genomics 2021; 7:000734 Available from: https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000734"
},
{
"DOI": "10.1016/j.meegid.2023.105480",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.7",
"unstructured": "Liang J , Ding Z , Liu K : Identification of critical SARS-CoV-2 amino acids associated with COVID-19 hospitalization rate using machine learning and statistical modeling: An observational study in the United States [Internet]. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases 2023; 113 Available from: https://pubmed.ncbi.nlm.nih.gov/37437768/"
},
{
"DOI": "10.21203/rs.3.rs-1234007/v1",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.8",
"unstructured": "Sokhansanj BA , Zhao Z , Rosen GL : An interpretable deep learning model for predicting the risk of severe COVID-19 from spike protein sequence [Internet] 2022; Available from: https://www.researchsquare.com"
},
{
"DOI": "10.1021/acs.jpclett.0c02765",
"article-title": "Decoding asymptomatic COVID-19 infection and transmission [Internet]",
"doi-asserted-by": "crossref",
"first-page": "10007",
"journal-title": "Journal of Physical Chemistry Letters",
"key": "2024031300400911000_2024.03.08.24303818v1.9",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.3389/fcimb.2022.868414",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.10",
"unstructured": "Maurya R , Mishra P , Swaminathan A , et al.: SARS-CoV-2 mutations and COVID-19 clinical outcome: Mutation global frequency dynamics and structural modulation hold the key [Internet]. Frontiers in cellular and infection microbiology 2022; 12 Available from: https://pubmed.ncbi.nlm.nih.gov/35386683/"
},
{
"DOI": "10.1016/j.meegid.2021.104730",
"article-title": "Different SARS-CoV-2 haplotypes associate with geographic origin and case fatality rates of COVID-19 patients",
"doi-asserted-by": "crossref",
"first-page": "104730",
"journal-title": "Infection, Genetics and Evolution",
"key": "2024031300400911000_2024.03.08.24303818v1.11",
"volume": "90",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2355-0",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.12"
},
{
"DOI": "10.3390/pathogens10091109",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.13",
"unstructured": "Mehta P , Alle S , Chaturvedi A , et al.: Clinico-genomic analysis reveals mutations associated with COVID-19 disease severity: Possible modulation by RNA structure [Internet]. Pathogens 2021, Vol 10, Page 1109 2021; 10:1109 Available from: https://www.mdpi.com/2076-0817/10/9/1109/htm"
},
{
"DOI": "10.1371/journal.pone.0271381",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.14",
"unstructured": "Zhu L , Marsh JW , Griffith MP , et al.: Predictive model for severe COVID-19 using SARS-CoV-2 whole-genome sequencing and electronic health record data, March 2020-may 2021 [Internet]. PloS one 2022; 17 Available from: https://pubmed.ncbi.nlm.nih.gov/35819967/"
},
{
"DOI": "10.1101/2022.08.13.22278740",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.15",
"unstructured": "Koch EM , Du J , Dressner M , et al.: Demographic and viral-genetic analyses of COVID- 19 severity in Bahrain identify local risk factors and a protective effect of polymerase mutations [Internet]. medRxiv : the preprint server for health sciences 2023; Available from: https://pubmed.ncbi.nlm.nih.gov/36032980/"
},
{
"DOI": "10.1016/j.csbj.2022.06.005",
"article-title": "Data-driven platform for identifying variants of interest in COVID-19 virus [Internet]",
"doi-asserted-by": "crossref",
"first-page": "2942",
"journal-title": "Computational and structural biotechnology journal",
"key": "2024031300400911000_2024.03.08.24303818v1.16",
"volume": "20",
"year": "2022"
},
{
"DOI": "10.1101/2022.04.15.22273922",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.17",
"unstructured": "Agarwal R , Leblond T , Mcauley EM , et al.: Linking genotype to phenotype: Further exploration of mutations in SARS-CoV-2 associated with mild or severe outcomes [Internet]. medRxiv 2022; 2022.04.15.22273922 Available from: https://www.medrxiv.org/content/10.1101/2022.04.15.22273922v1"
},
{
"DOI": "10.46234/ccdcw2021.255",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.18"
},
{
"DOI": "10.1145/2939672.2939785",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.19",
"unstructured": "Chen T , Guestrin C : XGBoost: A scalable tree boosting system. Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2016; 13–17-August-2016:785–794"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.20",
"unstructured": "Understand your dataset with XGBoost [Internet] Available from: https://cran.r-project.org/web/packages/xgboost/vignettes/discoverYourData.html#numeric-v.s.-categorical-variables"
},
{
"DOI": "10.1186/s12859-021-04034-6",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.21"
},
{
"DOI": "10.1126/science.abm1208",
"article-title": "Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness [Internet]",
"doi-asserted-by": "crossref",
"first-page": "1327",
"journal-title": "Science",
"key": "2024031300400911000_2024.03.08.24303818v1.22",
"volume": "376",
"year": "2022"
},
{
"DOI": "10.1001/jamanetworkopen.2023.9694",
"article-title": "Neutralizing monoclonal antibody use and COVID-19 infection outcomes [Internet]",
"doi-asserted-by": "crossref",
"first-page": "e239694",
"journal-title": "JAMA Network Open",
"key": "2024031300400911000_2024.03.08.24303818v1.23",
"volume": "6",
"year": "2023"
},
{
"DOI": "10.1101/2023.05.03.23289106",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.24",
"unstructured": "Ambrose N , B Amin A , Anderson B , et al.: Descriptive analysis of SARS-CoV-2 genomics data from ambulatory patients [Internet]. medRxiv 2023; 2023.05.03.23289106 Available from: https://www.medrxiv.org/content/10.1101/2023.05.03.23289106v1"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.25",
"unstructured": "Coronavirus (COVID-19) | drugs | FDA [Internet] Available from: https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs"
},
{
"DOI": "10.1097/00005650-199801000-00004",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.26"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.27",
"unstructured": "Neighborhood atlas - home [Internet] Available from: https://www.neighborhoodatlas.medicine.wisc.edu/"
},
{
"DOI": "10.1056/NEJMp1802313",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.28"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.29",
"unstructured": "Scikit-learn: Machine learning in python — scikit-learn 1.3.1 documentation [Internet] Available from: https://scikit-learn.org/stable/"
},
{
"DOI": "10.1038/s41588-021-00862-7",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.30"
},
{
"DOI": "10.1093/ve/veab064",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.31",
"unstructured": "O’Toole Á , Scher E , Underwood A , et al.: Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool [Internet]. Virus Evolution 2021; 7 Available from: https://academic.oup.com/ve/article/7/2/veab064/6315289"
},
{
"DOI": "10.21105/joss.03773",
"article-title": "Nextclade: Clade assignment, mutation calling and quality control for viral genomes [Internet]",
"doi-asserted-by": "crossref",
"first-page": "3773",
"journal-title": "Journal of Open Source Software",
"key": "2024031300400911000_2024.03.08.24303818v1.32",
"volume": "6",
"year": "2021"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.33",
"unstructured": "Package ’xgboost’ type package title extreme gradient boosting [Internet] 2023; Available from: https://github.com/dmlc/xgboost/issues"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.34",
"unstructured": "Yan Y : Machine learning evaluation metrics [r package MLmetrics version 1.1.1] [Internet] 2016; Available from: https://CRAN.R-project.org/package=MLmetrics"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.35",
"unstructured": "Lundberg SM , Allen PG , Lee S-I: A unified approach to interpreting model predictions [Internet]. Advances in Neural Information Processing Systems 2017; 30 Available from: https://github.com/slundberg/shap"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.36",
"unstructured": "Mayer M : SHAP visualizations [r package shapviz version 0.9.2] [Internet] 2023; Available from: https://CRAN.R-project.org/package=shapviz"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.37",
"unstructured": "Create elegant data visualisations using the grammar of graphics • ggplot2 [Internet] Available from: https://ggplot2.tidyverse.org/"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.38",
"unstructured": "Wingate D , Weber T : Automated variational inference in probabilistic programming [Internet] 2013; Available from: https://arxiv.org/abs/1301.1299v1"
},
{
"DOI": "10.48550/arXiv.1805.12177",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.39"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.40",
"unstructured": "Pyro documentation — pyro documentation [Internet] Available from: https://docs.pyro.ai/en/stable/index.html"
},
{
"DOI": "10.2307/2291073",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.41"
},
{
"DOI": "10.3758/PBR.16.2.225",
"article-title": "Bayesian t tests for accepting and rejecting the null hypothesis [Internet]",
"doi-asserted-by": "crossref",
"first-page": "225",
"journal-title": "Psychonomic Bulletin and Review",
"key": "2024031300400911000_2024.03.08.24303818v1.42",
"volume": "16",
"year": "2009"
},
{
"DOI": "10.1093/nar/28.1.235",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.43"
},
{
"article-title": "Highly accurate protein structure prediction with AlphaFold [Internet]",
"first-page": "7873",
"journal-title": "Nature",
"key": "2024031300400911000_2024.03.08.24303818v1.44",
"volume": "596",
"year": "2021"
},
{
"DOI": "10.1002/pro.4792",
"article-title": "UCSF ChimeraX: Tools for structure building and analysis [Internet]",
"doi-asserted-by": "crossref",
"first-page": "e4792",
"journal-title": "Protein Science",
"key": "2024031300400911000_2024.03.08.24303818v1.45",
"volume": "32",
"year": "2023"
},
{
"DOI": "10.1002/prot.22488",
"article-title": "Improved prediction of protein side-chain conformations with SCWRL4 [Internet]",
"doi-asserted-by": "crossref",
"first-page": "778",
"journal-title": "Proteins: Structure, Function, and Bioinformatics",
"key": "2024031300400911000_2024.03.08.24303818v1.46",
"volume": "77",
"year": "2009"
},
{
"DOI": "10.1126/scitranslmed.abk3445",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.47",
"unstructured": "Maher MC , Bartha I , Weaver S , et al.: Predicting the mutational drivers of future SARS-CoV-2 variants of concern [Internet]. Science translational medicine 2022; 14 Available from: https://pubmed.ncbi.nlm.nih.gov/35014856/"
},
{
"DOI": "10.1101/2021.09.12.21263475",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.48",
"unstructured": "Shen L , Triche TJ , Bard JD , et al.: Spike protein NTD mutation G142D in SARS-CoV-2 delta VOC lineages is associated with frequent back mutations, increased viral loads, and immune evasion [Internet]. medRxiv 2021; 2021.09.12.21263475 Available from: https://www.medrxiv.org/content/10.1101/2021.09.12.21263475v1"
},
{
"DOI": "10.1073/pnas.2101161118",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.49"
},
{
"DOI": "10.1371/journal.pcbi.1010963",
"article-title": "Inferring feature importance with uncertainties with application to large genotype data [Internet]",
"doi-asserted-by": "crossref",
"first-page": "e1010963",
"journal-title": "PLOS Computational Biology",
"key": "2024031300400911000_2024.03.08.24303818v1.50",
"volume": "19",
"year": "2023"
},
{
"DOI": "10.1186/s13059-018-1618-7",
"doi-asserted-by": "crossref",
"key": "2024031300400911000_2024.03.08.24303818v1.51",
"unstructured": "Grubaugh ND , Gangavarapu K , Quick J , et al.: An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar [Internet]. bioRxiv 2018; 383513 Available from: https://www.biorxiv.org/content/10.1101/383513v1"
},
{
"DOI": "10.4161/fly.19695",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.52"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.53",
"unstructured": "Index of /goldenPath/wuhCor1/UShER_SARS-CoV-2 [Internet] Available from: http://hgdownload.soe.ucsc.edu/goldenPath/wuhCor1/UShER_SARS-CoV-2/"
},
{
"key": "2024031300400911000_2024.03.08.24303818v1.54",
"unstructured": "United states - covSPECTRUM [Internet] Available from: https://cov-spectrum.org/explore/United%20States/AllSamples/Past6M"
},
{
"DOI": "10.1038/s41586-020-2008-3",
"doi-asserted-by": "publisher",
"key": "2024031300400911000_2024.03.08.24303818v1.55"
}
],
"reference-count": 55,
"references-count": 55,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2024.03.08.24303818"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "SARS-CoV-2 Genetic Variants and Patient Factors Associated with Hospitalization Risk",
"type": "posted-content"
}