SARS-CoV-2 Assembly: Gaining Infectivity and Beyond
Harshita Katiyar, Ariana Arduini, Yichen Li, Chen Liang
Viruses, doi:10.3390/v16111648
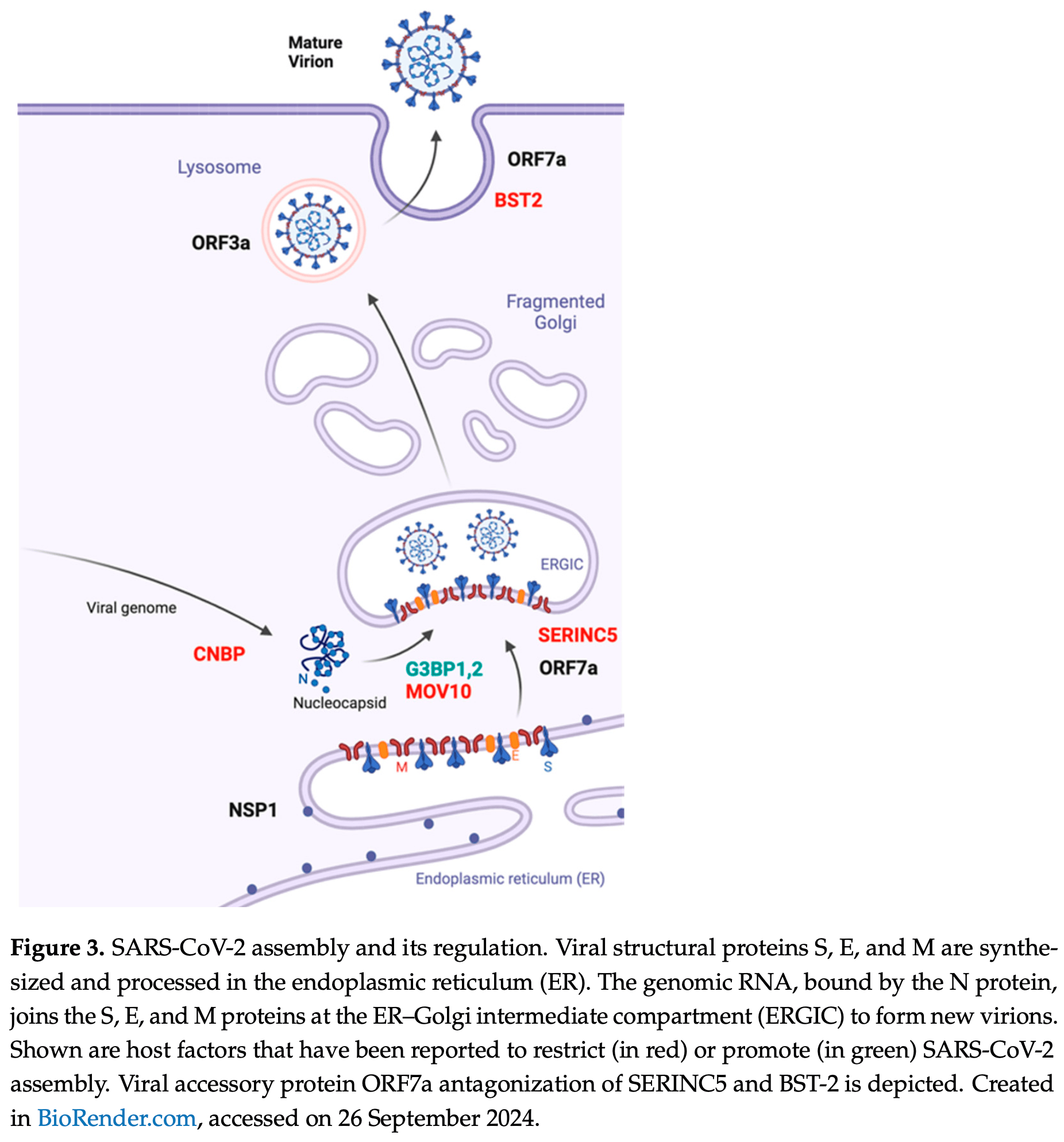
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was responsible for causing the COVID-19 pandemic. Intensive research has illuminated the complex biology of SARS-CoV-2 and its continuous evolution during and after the COVID-19 pandemic. While much attention has been paid to the structure and functions of the viral spike protein and the entry step of viral infection, partly because these are targets for neutralizing antibodies and COVID-19 vaccines, the later stages of SARS-CoV-2 replication, including the assembly and egress of viral progenies, remain poorly characterized. This includes insight into how the activities of the viral structural proteins are orchestrated spatially and temporally, which cellular proteins are assimilated by the virus to assist viral assembly, and how SARS-CoV-2 counters and evades the cellular mechanisms antagonizing virus assembly. In addition to becoming infectious, SARS-CoV-2 progenies also need to survive the hostile innate and adaptive immune mechanisms, such as recognition by neutralizing antibodies. This review offers an updated summary of the roles of SARS-CoV-2 structural proteins in viral assembly, the regulation of assembly by viral and cellular factors, and the cellular mechanisms that restrict this process. Knowledge of these key events often reveals the vulnerabilities of SARS-CoV-2 and aids in the development of effective antiviral therapeutics.
Author Contributions: Writing-original draft preparation, H.K. and Y.L.; writing-review and editing, H.K., A.A. and C.L.; visualization-figure preparation, H.K. All authors have read and agreed to the published version of the manuscript.
References
Abavisani, Rahimian, Mahdavi, Tokhanbigli, Mollapour Siasakht et al., Mutations in SARS-CoV-2 Structural Proteins: A Global Analysis, Virol. J,
doi:10.1186/s12985-022-01951-7Aldaais, Yegnaswamy, Albahrani, Alsowaiket, Alramadan, Sequence and Structural Analysis of COVID-19 E and M Proteins with MERS Virus E and M Proteins-A Comparative Study, Biochem. Biophys. Rep,
doi:10.1016/j.bbrep.2021.101023Ali, Prasad, Alasmari, Alharbi, Rashid et al., Genomics-Guided Targeting of Stress Granule Proteins G3BP1/2 to Inhibit SARS-CoV-2 Propagation, Int. J. Biol. Macromol,
doi:10.1016/j.ijbiomac.2021.09.018Alsaadi, Neuman, Jones, Identification of a Membrane Binding Peptide in the Envelope Protein of MHV Coronavirus, Viruses,
doi:10.3390/v12091054Arndt, Larson, Hogue, A Conserved Domain in the Coronavirus Membrane Protein Tail Is Important for Virus Assembly, J. Virol,
doi:10.1128/JVI.01131-10Baggen, Jacquemyn, Persoons, Vanstreels, Pye et al., TMEM106B Is a Receptor Mediating ACE2-Independent SARS-CoV-2 Cell Entry, Cell,
doi:10.1016/j.cell.2023.06.005Bayati, Kumar, Francis, Mcpherson, SARS-CoV-2 Infects Cells after Viral Entry via Clathrin-Mediated Endocytosis, J. Biol. Chem,
doi:10.1016/j.jbc.2021.100306Bergner, Zech, Hirschenberger, Stenger, Sparrer et al., Near-Native Visualization of SARS-CoV-2 Induced Membrane Remodeling and Virion Morphogenesis, Viruses,
doi:10.3390/v14122786Beske, Reichelt, Taylor, Kirkegaard, Andino, Poliovirus Infection Blocks ERGIC-to-Golgi Trafficking and Induces Microtubule-Dependent Disruption of the Golgi Complex, J. Cell Sci,
doi:10.1242/jcs.03483Bianchi, Benvenuto, Giovanetti, Angeletti, Ciccozzi et al., Sars-CoV-2 Envelope and Membrane Proteins: Structural Differences Linked to Virus Characteristics?, BioMed Res. Int,
doi:10.1155/2020/4389089Boson, Legros, Zhou, Siret, Mathieu et al., The SARS-CoV-2 Envelope and Membrane Proteins Modulate Maturation and Retention of the Spike Protein, Allowing Assembly of Virus-like Particles, J. Biol. Chem,
doi:10.1074/jbc.RA120.016175Botova, Camacho-Zarco, Tognetti, Bessa, Guseva et al., A Specific Phosphorylation-Dependent Conformational Switch in SARS-CoV-2 Nucleocapsid Protein Inhibits RNA Binding, Sci. Adv,
doi:10.1126/sciadv.aax2323Bracquemond, Muriaux, Betacoronavirus Assembly: Clues and Perspectives for Elucidating SARS-CoV-2 Particle Formation and Egress, mBio,
doi:10.1128/mBio.02371-21Brun, Vasiljevic, Gangadharan, Hensen, Chandran et al., Assessing Antigen Structural Integrity through Glycosylation Analysis of the SARS-CoV-2 Viral Spike, ACS Cent. Sci,
doi:10.1021/acscentsci.1c00058Cai, Yu, Wang, Liang, Richard, Arginine Methylation of SARS-CoV-2 Nucleocapsid Protein Regulates RNA Binding, Its Ability to Suppress Stress Granule Formation, and Viral Replication, J. Biol. Chem,
doi:10.1016/j.jbc.2021.100821Carabelli, Peacock, Thorne, Harvey, Hughes, COVID-19
Carlson, Adly, Bi, Howard, Frost et al., Reconstitution of the SARS-CoV-2 Ribonucleosome Provides Insights into Genomic RNA Packaging and Regulation by Phosphorylation, J. Biol. Chem,
doi:10.1016/j.jbc.2022.102560Castro, Pérez-Berna, Calvo, Pereiro, Gastaminza, Three-Dimensional Remodeling of SARS-CoV2-Infected Cells Revealed by Cryogenic Soft X-Ray Tomography, ACS Nano,
doi:10.1021/acsnano.3c07265Cattin-Ortolá, Welch, Maslen, Papa, James et al., Sequences in the Cytoplasmic Tail of SARS-CoV-2 Spike Facilitate Expression at the Cell Surface and Syncytia Formation, Nat. Commun,
doi:10.1038/s41467-021-25589-1Chang, Chu, Chen, Wu, Su et al., Development of Fluorescence-Tagged SARS-CoV-2 Virus-like Particles by a Tri-Cistronic Vector Expression System for Investigating the Cellular Entry of SARS-CoV-2, Viruses,
doi:10.3390/v14122825Chen, Lei, Jiang, Humphries, Parsi et al., Cellular Nucleic Acid-Binding Protein Restricts SARS-CoV-2 by Regulating Interferon and Disrupting RNA-Protein Condensates, Proc. Natl. Acad. Sci,
doi:10.1073/pnas.2308355120Cole, Sciaky, Marotta, Song, Lippincott-Schwartz, Golgi Dispersal during Microtubule Disruption: Regeneration of Golgi Stacks at Peripheral Endoplasmic Reticulum Exit Sites, Mol. Biol. Cell,
doi:10.1091/mbc.7.4.631Collins, Steer, Binding of the SARS-CoV-2 Spike Protein to the Asialoglycoprotein Receptor on Human Primary Hepatocytes and Immortalized Hepatocyte-Like Cells by Confocal Analysis, Hepatic Med. Evid. Res,
doi:10.2147/HMER.S301979Cortese, Lee, Cerikan, Neufeldt, Oorschot et al., Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies, Cell Host Microbe,
doi:10.1016/j.chom.2020.11.003Cubuk, Alston, Incicco, Singh, Stuchell-Brereton et al., The SARS-CoV-2 Nucleocapsid Protein Is Dynamic, Disordered, and Phase Separates with RNA, Nat. Commun,
doi:10.1038/s41467-021-21953-3De Haan, Rottier, Molecular Interactions in the Assembly of Coronaviruses
De Haan, Smeets, Vernooij, Vennema, Rottier, Mapping of the Coronavirus Membrane Protein Domains Involved in Interaction with the Spike Protein, J. Virol,
doi:10.1128/JVI.73.9.7441-7452.1999De Silva, Peacock, Barclay, De Silva, SARS-CoV-2 Variant Biology: Immune Escape, Transmission and Fitness, Nat. Rev. Microbiol,
doi:10.1038/s41579-022-00841-7Demirov, Ono, Orenstein, Freed, Overexpression of the N-Terminal Domain of TSG101 Inhibits HIV-1 Budding by Blocking Late Domain Function, Proc. Natl. Acad. Sci,
doi:10.1128/JVI.76.10.4679-4687.2002Desmarets, Danneels, Burlaud-Gaillard, Blanchard, Dubuisson et al., The KxGxYR and DxE Motifs in the C-Tail of the Middle East Respiratory Syndrome Coronavirus Membrane Protein Are Crucial for Infectious Virus Assembly, Cell. Mol. Life Sci. CMLS,
doi:10.1007/s00018-023-05008-yDu, Deiter, Bouzidi, Billaud, Simmons et al., A Viral Assembly Inhibitor Blocks SARS-CoV-2 Replication in Airway Epithelial Cells, Commun. Biol,
doi:10.1038/s42003-024-06130-8Duart, García-Murria, Grau, Acosta-Cáceres, Martínez-Gil et al., SARS-CoV-2 Envelope Protein Topology in Eukaryotic Membranes, Open Biol,
doi:10.1098/rsob.200209Ewart, Bobardt, Bentzen, Yan, Thomson et al., Post-Infection Treatment with the E Protein Inhibitor BIT225 Reduces Disease Severity and Increases Survival of K18-hACE2 Transgenic Mice Infected with a Lethal Dose of SARS-CoV-2, PLoS Pathog,
doi:10.1371/journal.ppat.1011328Finkel, Gluck, Nachshon, Winkler, Fisher et al., SARS-CoV-2 Uses a Multipronged Strategy to Impede Host Protein Synthesis, Nature,
doi:10.1038/s41586-021-03610-3Gallop, Jao, Kent, Butler, Evans et al., Mechanism of Endophilin N-BAR Domain-Mediated Membrane Curvature, EMBO J,
doi:10.1038/sj.emboj.7601174Garrus, Von Schwedler, Pornillos, Morham, Zavitz et al., Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding, Cell,
doi:10.1016/S0092-8674(01)00506-2Ghosh, Dellibovi-Ragheb, Kerviel, Pak, Qiu et al., β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway, Cell,
doi:10.1016/j.cell.2020.10.039Gourdelier, Swain, Arone, Mouttou, Bracquemond et al., Optimized Production and Fluorescent Labeling of SARS-CoV-2 Virus-like Particles, Sci. Rep,
doi:10.1038/s41598-022-18681-zGu, Cao, Zhang, Gao, Wang et al., Receptome Profiling Identifies KREMEN1 and ASGR1 as Alternative Functional Receptors of SARS-CoV-2, Cell Res,
doi:10.1038/s41422-021-00595-6Harvey, Carabelli, Jackson, Gupta, Thomson et al., COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape, Nat. Rev. Microbiol,
doi:10.1038/s41579-021-00573-0Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell,
doi:10.1016/j.cell.2020.02.052Hsieh, Chang, Huang, Lee, Hsiao et al., Assembly of Severe Acute Respiratory Syndrome Coronavirus RNA Packaging Signal into Virus-Like Particles Is Nucleocapsid Dependent, J. Virol,
doi:10.1128/JVI.79.22.13848-13855.2005Hu, Guo, Zhou, Shi, Characteristics of SARS-CoV-2 and COVID-19, Nat. Rev. Microbiol
Hu, Siche, Möller, Veit, Amphipathic Helices of Cellular Proteins Can Replace the Helix in M2 of Influenza A Virus with Only Small Effects on Virus Replication, J. Virol,
doi:10.1128/JVI.01605-19Huang, Wang, Zhong, Zhang, Zhang et al., Molecular Architecture of Coronavirus Double-Membrane Vesicle Pore Complex, Nature,
doi:10.1038/s41586-024-07817-yJennings, Kornfeld, Doray, A Weak COPI Binding Motif in the Cytoplasmic Tail of SARS-CoV-2 Spike Glycoprotein Is Necessary for Its Cleavage, Glycosylation, and Localization, FEBS Lett,
doi:10.1002/1873-3468.14109Khan, Li, Tao, Wang, Ye et al., Tubeimosides Are Pan-Coronavirus and Filovirus Inhibitors That Can Block Their Fusion Protein Binding to Niemann-Pick C1, Nat. Commun,
doi:10.1038/s41467-023-44504-4Klein, Cortese, Winter, Wachsmuth-Melm, Neufeldt et al., SARS-CoV-2 Structure and Replication Characterized by in Situ Cryo-Electron Tomography, Nat. Commun,
doi:10.1038/s41467-020-19619-7Kumar, Hawkins, Kicmal, Qing, Timm et al., Assembly and Entry of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV2): Evaluation Using Virus-Like Particles, Cells,
doi:10.3390/cells10040853Kumar, Kumar, Garg, Giri, An Insight into SARS-CoV-2 Membrane Protein Interaction with Spike, Envelope, and Nucleocapsid Proteins, J. Biomol. Struct. Dyn,
doi:10.1080/07391102.2021.2016490Kuzmin, Orekhov, Astashkin, Gordeliy, Gushchin, Structure and Dynamics of the SARS-CoV-2 Envelope Protein Monomer, Proteins,
doi:10.1002/prot.26317Liao, Wang, Tang, Yang, Duan et al., Human Transferrin Receptor Can Mediate SARS-CoV-2 Infection, Proc. Natl. Acad. Sci,
doi:10.1073/pnas.2317026121Liu, Huuskonen, Laitinen, Redchuk, Bogacheva et al., SARS-CoV-2-Host Proteome Interactions for Antiviral Drug Discovery, Mol. Syst. Biol,
doi:10.15252/msb.202110396Lu, Liu, Zhao, Gomez Castro, Laurent-Rolle et al., SARS-CoV-2 Exacerbates Proinflammatory Responses in Myeloid Cells through C-Type Lectin Receptors and Tweety Family Member 2, Immunity,
doi:10.1016/j.immuni.2021.05.006Lu, Ye, Singh, Cao, Diedrich et al., The SARS-CoV-2 Nucleocapsid Phosphoprotein Forms Mutually Exclusive Condensates with RNA and the Membrane-Associated M Protein, Nat. Commun,
doi:10.1038/s41467-020-20768-yLusvarghi, Stauft, Vassell, Williams, Baha et al., Effects of N-Glycan Modifications on Spike Expression, Virus Infectivity, and Neutralization Sensitivity in Ancestral Compared to Omicron SARS-CoV-2 Variants, PLoS Pathog,
doi:10.1371/journal.ppat.1011788Maeda, Maeda, Makino, Release of Coronavirus E Protein in Membrane Vesicles from Virus-Infected Cells and E Protein-Expressing Cells, Virology,
doi:10.1006/viro.1999.9955Mandala, Mckay, Shcherbakov, Dregni, Kolocouris et al., Structure and Drug Binding of the SARS-CoV-2 Envelope Protein Transmembrane Domain in Lipid Bilayers, Nat. Struct. Mol. Biol,
doi:10.1038/s41594-020-00536-8Martin-Serrano, Zang, Bieniasz, HIV-1 and Ebola Virus Encode Small Peptide Motifs That Recruit Tsg101 to Sites of Particle Assembly to Facilitate Egress, Nat. Med,
doi:10.1038/nm1201-1313Martyna, Bahsoun, Badham, Srinivasan, Howard et al., Membrane Remodeling by the M2 Amphipathic Helix Drives Influenza Virus Membrane Scission, Sci. Rep,
doi:10.1038/srep44695Masters, The Molecular Biology of Coronaviruses
Mcbride, Li, Machamer, The Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Contains a Novel Endoplasmic Reticulum Retrieval Signal That Binds COPI and Promotes Interaction with Membrane Protein, J. Virol,
doi:10.1128/JVI.02146-06Medeiros-Silva, Dregni, Somberg, Duan, Hong, Atomic Structure of the Open SARS-CoV-2 E Viroporin, Sci. Adv,
doi:10.1126/sciadv.adi9007Mendonça, Howe, Gilchrist, Sheng, Sun et al., Correlative Multi-Scale Cryo-Imaging Unveils SARS-CoV-2 Assembly and Egress, Nat. Commun,
doi:10.1038/s41467-021-24887-yMeng, Ip, Prestwood, Abbink, Lever, Evidence That the Endosomal Sorting Complex Required for Transport-II (ESCRT-II) Is Required for Efficient Human Immunodeficiency Virus-1 (HIV-1) Production, Retrovirology,
doi:10.1186/s12977-015-0197-xMiura, Suzuki, Ishida, Arakawa, Wu et al., Distinct Motifs in the E Protein Are Required for SARS-CoV-2 Virus Particle Formation and Lysosomal Deacidification in Host Cells, J. Virol,
doi:10.1128/jvi.00426-23Mohan, Wollert, Membrane Remodeling by SARS-CoV-2 -Double-Enveloped Viral Replication, Fac. Rev,
doi:10.12703/r/10-17Monje-Galvan, Voth, Molecular Interactions of the M and E Integral Membrane Proteins of SARS-CoV-2, Faraday Discuss,
doi:10.1039/D1FD00031DMoradi, Wu, Walden, Cui, Johnston et al., In Vitro Reconstitution and Analysis of SARS-CoV-2/Host Protein-Protein Interactions, ACS Omega,
doi:10.1021/acsomega.3c01625Murigneux, Softic, Aubé, Grandi, Judith et al., Proteomic Analysis of SARS-CoV-2 Particles Unveils a Key Role of G3BP Proteins in Viral Assembly, Nat. Commun,
doi:10.1038/s41467-024-44958-0Owen, Allerton, Anderson, Aschenbrenner, Avery et al., An Oral SARS-CoV-2 M pro Inhibitor Clinical Candidate for the Treatment of COVID-19, Science,
doi:10.1126/science.abl4784Pearson, Mears, Broncel, Snijders, Bauer et al., ER-Export and ARFRP1/AP-1-Dependent Delivery of SARS-CoV-2 Envelope to Lysosomes Controls Late Stages of Viral Replication, Sci. Adv,
doi:10.1126/sciadv.adl5012Peng, Du, Lei, Dorje, Qi et al., Structures of the SARS -CoV-2 Nucleocapsid and Their Perspectives for Drug Design, EMBO J,
doi:10.15252/embj.2020105938Perez-Miller, Patek, Moutal, Duran, Cabel et al., Novel Compounds Targeting Neuropilin Receptor 1 with Potential To Interfere with SARS-CoV-2 Virus Entry, ACS Chem. Neurosci,
doi:10.1021/acschemneuro.0c00619Plescia, David, Patra, Sengupta, Amiar et al., SARS-CoV-2 Viral Budding and Entry Can Be Modeled Using BSL-2 Level Virus-like Particles, J. Biol. Chem,
doi:10.1074/jbc.RA120.016148Puthenveetil, Lun, Murphy, Healy, Vilmen et al., S-Acylation of SARS-CoV-2 Spike Protein: Mechanistic Dissection, in Vitro Reconstitution and Role in Viral Infectivity, J. Biol. Chem,
doi:10.1016/j.jbc.2021.101112Ricciardi, Guarino, Giaquinto, Polishchuk, Santoro et al., The Role of NSP6 in the Biogenesis of the SARS-CoV-2 Replication Organelle, Nature,
doi:10.1038/s41586-022-04835-6Rogalski, Bergmann, Singer, Effect of Microtubule Assembly Status on the Intracellular Processing and Surface Expression of an Integral Protein of the Plasma Membrane, J. Cell Biol,
doi:10.1083/jcb.99.3.1101Roingeard, Eymieux, Burlaud-Gaillard, Hourioux, Patient et al., The Double-Membrane Vesicle (DMV): A Virus-Induced Organelle Dedicated to the Replication of SARS-CoV-2 and Other Positive-Sense Single-Stranded RNA Viruses, Cell. Mol. Life Sci. CMLS,
doi:10.1007/s00018-022-04469-xSavastano, Ibáñez De Opakua, Rankovic, Zweckstetter, Nucleocapsid Protein of SARS-CoV-2 Phase Separates into RNA-Rich Polymerase-Containing Condensates, Nat. Commun,
doi:10.1038/s41467-020-19843-1Scherer, Mascheroni, Carnell, Wunderlich, Makarchuk et al., SARS-CoV-2 Nucleocapsid Protein Adheres to Replication Organelles before Viral Assembly at the Golgi/ERGIC and Lysosome-Mediated Egress, Sci. Adv,
doi:10.1126/sciadv.abl4895Schmidt, Mishra, Wang, Degrado, Wong, Influenza Virus A M2 Protein Generates Negative Gaussian Membrane Curvature Necessary for Budding and Scission, J. Am. Chem. Soc,
doi:10.1021/ja400146zSchubert, Karousis, Jomaa, Scaiola, Echeverria et al., SARS-CoV-2 Nsp1 Binds the Ribosomal mRNA Channel to Inhibit Translation, Nat. Struct. Mol. Biol,
doi:10.1038/s41594-020-0511-8Schöneberg, Lee, Iwasa, Hurley, Reverse-Topology Membrane Scission by the ESCRT Proteins, Nat. Rev. Mol. Cell Biol,
doi:10.1038/nrm.2016.121Sheahan, Sims, Zhou, Graham, Pruijssers et al., An Orally Bioavailable Broad-Spectrum Antiviral Inhibits SARS-CoV-2 in Human Airway Epithelial Cell Cultures and Multiple Coronaviruses in Mice, Sci. Transl. Med,
doi:10.1126/scitranslmed.abb5883Siu, Teoh, Lo, Chan, Kien et al., Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus Are Required for Efficient Assembly, Trafficking, and Release of Virus-Like Particles, J. Virol,
doi:10.1128/JVI.01052-08Snijder, Limpens, De Wilde, Jong, Zevenhoven-Dobbe et al., A Unifying Structural and Functional Model of the Coronavirus Replication Organelle: Tracking down RNA Synthesis, PLoS Biol,
doi:10.1371/journal.pbio.3000715Stukalov, Girault, Grass, Karayel, Bergant et al., Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV, Nature,
doi:10.1038/s41586-021-03493-4Stuwe, Reardon, Yu, Shah, Hughes et al., Phosphorylation in the Ser/Arg-Rich Region of the Nucleocapsid of SARS-CoV-2 Regulates Phase Separation by Inhibiting Self-Association of a Distant Helix, J. Biol. Chem,
doi:10.1016/j.jbc.2024.107354Sun, Karki, Aguilera, Lopez Hernandez, Sun et al., Computational Study on the Function of Palmitoylation on the Envelope Protein in SARS-CoV-2, J. Chem. Theory Comput,
doi:10.1021/acs.jctc.1c00359Sun, Qie, Liu, Ren, Li et al., Clinical Characteristics of Hospitalized Patients with SARS-CoV-2 Infection: A Single Arm Meta-Analysis, J. Med. Virol,
doi:10.1002/jmv.25735Sun, Zheng, Ji, Zhou, Su et al., Mass Spectrometry Analysis of SARS-CoV-2 Nucleocapsid Protein Reveals Camouflaging Glycans and Unique Post-Translational Modifications, Infect. Microbes Dis,
doi:10.1097/IM9.0000000000000071Syed, Taha, Tabata, Chen, Ciling et al., Rapid Assessment of SARS-CoV-2-Evolved Variants Using Virus-like Particles, Science,
doi:10.1126/science.abl6184Taylor, Coleman, Postel, Sisk, Bernbaum et al., Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference, J. Virol,
doi:10.1128/JVI.02274-15Thoms, Buschauer, Ameismeier, Koepke, Denk et al., Structural Basis for Translational Shutdown and Immune Evasion by the Nsp1 Protein of SARS-CoV-2, Science,
doi:10.1126/science.abc8665Tien, Tsai, Chen, Chou, Zhang et al., Glycosylation and S-Palmitoylation Regulate SARS-CoV-2 Spike Protein Intracellular Trafficking, iScience,
doi:10.1016/j.isci.2022.104709Timilsina, Umthong, Ivey, Waxman, Stavrou, SARS-CoV-2 ORF7a Potently Inhibits the Antiviral Effect of the Host Factor SERINC5, Nat. Commun,
doi:10.1038/s41467-022-30609-9V'kovski, Kratzel, Steiner, Stalder, Thiel, Coronavirus Biology and Replication: Implications for SARS-CoV-2, Nat. Rev. Microbiol,
doi:10.1038/s41579-020-00468-6Verdiá-Báguena, Nieto-Torres, Alcaraz, Dediego, Torres et al., Coronavirus E Protein Forms Ion Channels with Functionally and Structurally-Involved Membrane Lipids, Virology,
doi:10.1016/j.virol.2012.07.005Verplank, Bouamr, Lagrassa, Agresta, Kikonyogo et al., Tsg101, a Homologue of Ubiquitin-Conjugating (E2) Enzymes, Binds the L Domain in HIV Type 1 Pr55 Gag, Proc. Natl. Acad. Sci,
doi:10.1073/pnas.131059198Vora, Fontana, Mao, Leger, Zhang et al., Targeting Stem-Loop 1 of the SARS-CoV-2 5 ′ UTR to Suppress Viral Translation and Nsp1 Evasion, Proc. Natl. Acad. Sci,
doi:10.1073/pnas.2117198119Walls, Park, Tortorici, Wall, Mcguire et al., Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein, Cell,
doi:10.1016/j.cell.2020.02.058Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-nCoV) in Vitro, Cell Res,
doi:10.1038/s41422-020-0282-0Wang, Carreras-Sureda, Demaurex, SARS-CoV-2 Infection Alkalinizes the ERGIC and Lysosomes through the Viroporin Activity of the Viral Envelope Protein, J. Cell Sci,
doi:10.1242/jcs.260685Wang, Pan, Ji, Zuo, Xiao et al., Impact of SARS-CoV-2 Envelope Protein Mutations on the Pathogenicity of Omicron XBB, Cell Discov,
doi:10.1038/s41421-023-00575-7Wang, Sola, Enjuanes, Zuñiga, MOV10 Helicase Interacts with Coronavirus Nucleocapsid Protein and Has Antiviral Activity, mBio,
doi:10.1128/mBio.01316-21Welker, Paillart, Bernacchi, Importance of Viral Late Domains in Budding and Release of Enveloped RNA Viruses, Viruses,
doi:10.3390/v13081559Wolff, Limpens, Zevenhoven-Dobbe, Laugks, Zheng et al., A Molecular Pore Spans the Double Membrane of the Coronavirus Replication Organelle, Science,
doi:10.1126/science.abd3629Wrapp, Wang, Corbett, Goldsmith, Hsieh et al., Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation, Science,
doi:10.1126/science.abb2507Wu, Zhang, Wang, Zhang, Ren et al., Palmitoylation of SARS-CoV-2 S Protein Is Essential for Viral Infectivity, Signal Transduct. Target. Ther,
doi:10.1038/s41392-021-00651-yXia, Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design, Viruses,
doi:10.3390/v13010109Xu, Shi, Li, Song, Li, Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System, Front. Bioeng. Biotechnol,
doi:10.3389/fbioe.2020.00862Yan, Yang, Li, Zhang, Zheng et al., Coupling of N7-Methyltransferase and 3 ′ -5 ′ Exoribonuclease with SARS-CoV-2 Polymerase Reveals Mechanisms for Capping and Proofreading, Cell,
doi:10.1016/j.cell.2021.05.033Yan, Zhang, Ge, Zheng, Gao et al., Architecture of a SARS-CoV-2 Mini Replication and Transcription Complex, Nat. Commun,
doi:10.1038/s41467-020-19770-1Yuan, Hu, Xiao, Tan, Li et al., The E3 Ubiquitin Ligase RNF5 Facilitates SARS-CoV-2 Membrane Protein-Mediated Virion Release, mBio,
doi:10.1128/mbio.03168-21Zhang, Jackson, Mou, Ojha, Peng et al., SARS-CoV-2 Spike-Protein D614G Mutation Increases Virion Spike Density and Infectivity, Nat. Commun,
doi:10.1038/s41467-020-19808-4Zhang, Kennedy, De Melo Jorge, Xing, Reid et al., SARS-CoV-2 Remodels the Golgi Apparatus to Facilitate Viral Assembly and Secretion, BioRxiv Prepr. Serv. Biol,
doi:10.1101/2022.03.04.483074Zhang, Li, Cruz Cosme, Gerzanich, Tang et al., Genome-Wide Characterization of SARS-CoV-2 Cytopathogenic Proteins in the Search of Antiviral Targets, mBio,
doi:10.1128/mbio.00169-22Zhang, Li, Feng, Ramos Da Silva, Ju et al., SARS-CoV-2 Pseudovirus Infectivity and Expression of Viral Entry-related Factors ACE2, TMPRSS2, Kim-1, and NRP-1 in Human Cells from the Respiratory, Urinary, Digestive, Reproductive, and Immune Systems, J. Med. Virol,
doi:10.1002/jmv.27244Zhang, Nomura, Muramoto, Ekimoto, Uemura et al., Structure of SARS-CoV-2 Membrane Protein Essential for Virus Assembly, Nat. Commun,
doi:10.1038/s41467-022-32019-3Zhang, Schmidt, Muecksch, Wang, Gazumyan et al., SARS-CoV-2 Spike Glycosylation Affects Function and Neutralization Sensitivity, mBio,
doi:10.1128/mbio.01672-23Zhao, Praissman, Grant, Cai, Xiao et al., Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor, Cell Host Microbe,
doi:10.1016/j.chom.2020.08.004Zhou, Liu, Gupta, Paramo, Hou et al., A Comprehensive SARS-CoV-2-Human Protein-Protein Interactome Reveals COVID-19 Pathobiology and Potential Host Therapeutic Targets, Nat. Biotechnol,
doi:10.1038/s41587-022-01474-0Zhou, Yang, Wang, Hu, Zhang et al., A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin, Nature,
doi:10.1038/s41586-020-2012-7Zhu, Zhang, Wang, Li, Yang et al., A Novel Coronavirus from Patients with Pneumonia in China, N. Engl. J. Med,
doi:10.1056/NEJMoa2001017Zhukovsky, Filograna, Luini, Corda, Valente, Protein Amphipathic Helix Insertion: A Mechanism to Induce Membrane Fission, Front. Cell Dev. Biol,
doi:10.3389/fcell.2019.00291Zimmermann, Zhao, Makroczyova, Wachsmuth-Melm, Prasad et al., SARS-CoV-2 Nsp3 and Nsp4 Are Minimal Constituents of a Pore Spanning Replication Organelle, Nat. Commun,
doi:10.1038/s41467-023-43666-5DOI record:
{
"DOI": "10.3390/v16111648",
"ISSN": [
"1999-4915"
],
"URL": "http://dx.doi.org/10.3390/v16111648",
"abstract": "<jats:p>Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was responsible for causing the COVID-19 pandemic. Intensive research has illuminated the complex biology of SARS-CoV-2 and its continuous evolution during and after the COVID-19 pandemic. While much attention has been paid to the structure and functions of the viral spike protein and the entry step of viral infection, partly because these are targets for neutralizing antibodies and COVID-19 vaccines, the later stages of SARS-CoV-2 replication, including the assembly and egress of viral progenies, remain poorly characterized. This includes insight into how the activities of the viral structural proteins are orchestrated spatially and temporally, which cellular proteins are assimilated by the virus to assist viral assembly, and how SARS-CoV-2 counters and evades the cellular mechanisms antagonizing virus assembly. In addition to becoming infectious, SARS-CoV-2 progenies also need to survive the hostile innate and adaptive immune mechanisms, such as recognition by neutralizing antibodies. This review offers an updated summary of the roles of SARS-CoV-2 structural proteins in viral assembly, the regulation of assembly by viral and cellular factors, and the cellular mechanisms that restrict this process. Knowledge of these key events often reveals the vulnerabilities of SARS-CoV-2 and aids in the development of effective antiviral therapeutics.</jats:p>",
"alternative-id": [
"v16111648"
],
"author": [
{
"affiliation": [
{
"name": "Lady Davis Institute, Jewish General Hospital, Montreal, QC H3T 1E2, Canada"
},
{
"name": "Department of Microbiology and Immunology, McGill University, Montreal, QC H3A 2B4, Canada"
}
],
"family": "Katiyar",
"given": "Harshita",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Lady Davis Institute, Jewish General Hospital, Montreal, QC H3T 1E2, Canada"
},
{
"name": "Department of Medicine, McGill University, Montreal, QC H3G 2M1, Canada"
}
],
"family": "Arduini",
"given": "Ariana",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Lady Davis Institute, Jewish General Hospital, Montreal, QC H3T 1E2, Canada"
},
{
"name": "Department of Microbiology and Immunology, McGill University, Montreal, QC H3A 2B4, Canada"
}
],
"family": "Li",
"given": "Yichen",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Lady Davis Institute, Jewish General Hospital, Montreal, QC H3T 1E2, Canada"
},
{
"name": "Department of Microbiology and Immunology, McGill University, Montreal, QC H3A 2B4, Canada"
},
{
"name": "Department of Medicine, McGill University, Montreal, QC H3G 2M1, Canada"
}
],
"family": "Liang",
"given": "Chen",
"sequence": "additional"
}
],
"container-title": "Viruses",
"container-title-short": "Viruses",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
10,
22
]
],
"date-time": "2024-10-22T08:10:14Z",
"timestamp": 1729584614000
},
"deposited": {
"date-parts": [
[
2024,
10,
22
]
],
"date-time": "2024-10-22T08:33:33Z",
"timestamp": 1729586013000
},
"funder": [
{
"award": [
"PJT-166048",
"PJT-185996"
],
"name": "Canadian Institutes of Health Research"
}
],
"indexed": {
"date-parts": [
[
2024,
10,
23
]
],
"date-time": "2024-10-23T04:17:33Z",
"timestamp": 1729657053364,
"version": "3.28.0"
},
"is-referenced-by-count": 0,
"issue": "11",
"issued": {
"date-parts": [
[
2024,
10,
22
]
]
},
"journal-issue": {
"issue": "11",
"published-online": {
"date-parts": [
[
2024,
11
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
10,
22
]
],
"date-time": "2024-10-22T00:00:00Z",
"timestamp": 1729555200000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1999-4915/16/11/1648/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1648",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2024,
10,
22
]
]
},
"published-online": {
"date-parts": [
[
2024,
10,
22
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1056/NEJMoa2001017",
"article-title": "A Novel Coronavirus from Patients with Pneumonia in China, 2019",
"author": "Zhu",
"doi-asserted-by": "crossref",
"first-page": "727",
"journal-title": "N. Engl. J. Med.",
"key": "ref_1",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1038/s41564-020-0695-z",
"doi-asserted-by": "crossref",
"key": "ref_2",
"unstructured": "Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020). The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-nCoV and Naming It SARS-CoV-2. Nat. Microbiol., 5, 536–544."
},
{
"key": "ref_3",
"unstructured": "(2024, September 27). World Health Organization WHO COVID-19 Dashboard, Available online: https://data.who.int/dashboards/covid19/cases."
},
{
"DOI": "10.1038/s41579-020-00468-6",
"article-title": "Coronavirus Biology and Replication: Implications for SARS-CoV-2",
"author": "Kratzel",
"doi-asserted-by": "crossref",
"first-page": "155",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_4",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1038/s41579-020-00459-7",
"article-title": "Characteristics of SARS-CoV-2 and COVID-19",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "141",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_5",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1002/jmv.25735",
"article-title": "Clinical Characteristics of Hospitalized Patients with SARS-CoV-2 Infection: A Single Arm Meta-Analysis",
"author": "Sun",
"doi-asserted-by": "crossref",
"first-page": "612",
"journal-title": "J. Med. Virol.",
"key": "ref_6",
"volume": "92",
"year": "2020"
},
{
"DOI": "10.1056/NEJMcp2009249",
"article-title": "Mild or Moderate COVID-19",
"author": "Gandhi",
"doi-asserted-by": "crossref",
"first-page": "1757",
"journal-title": "N. Engl. J. Med.",
"key": "ref_7",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1056/NEJMcp2009575",
"article-title": "Severe COVID-19",
"author": "Berlin",
"doi-asserted-by": "crossref",
"first-page": "2451",
"journal-title": "N. Engl. J. Med.",
"key": "ref_8",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1038/s41579-021-00573-0",
"article-title": "SARS-CoV-2 Variants, Spike Mutations and Immune Escape",
"author": "Harvey",
"doi-asserted-by": "crossref",
"first-page": "409",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_9",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1038/s41579-022-00841-7",
"doi-asserted-by": "crossref",
"key": "ref_10",
"unstructured": "10. Carabelli, A.M., Peacock, T.P., Thorne, L.G., Harvey, W.T., Hughes, J., COVID-19 Genomics UK Consortium, De Silva, T.I., Peacock, S.J., Barclay, W.S., and De Silva, T.I. (2023). SARS-CoV-2 Variant Biology: Immune Escape, Transmission and Fitness. Nat. Rev. Microbiol., 21, 162–177."
},
{
"DOI": "10.1038/s41579-023-01003-z",
"article-title": "SARS-CoV-2 Biology and Host Interactions",
"author": "Steiner",
"doi-asserted-by": "crossref",
"first-page": "206",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_11",
"volume": "22",
"year": "2024"
},
{
"DOI": "10.1126/scitranslmed.abb5883",
"article-title": "An Orally Bioavailable Broad-Spectrum Antiviral Inhibits SARS-CoV-2 in Human Airway Epithelial Cell Cultures and Multiple Coronaviruses in Mice",
"author": "Sheahan",
"doi-asserted-by": "crossref",
"first-page": "eabb5883",
"journal-title": "Sci. Transl. Med.",
"key": "ref_12",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1126/science.abl4784",
"article-title": "An Oral SARS-CoV-2 M pro Inhibitor Clinical Candidate for the Treatment of COVID-19",
"author": "Owen",
"doi-asserted-by": "crossref",
"first-page": "1586",
"journal-title": "Science",
"key": "ref_13",
"volume": "374",
"year": "2021"
},
{
"DOI": "10.1038/s41422-020-0282-0",
"article-title": "Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-nCoV) in Vitro",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "269",
"journal-title": "Cell Res.",
"key": "ref_14",
"volume": "30",
"year": "2020"
},
{
"DOI": "10.1128/mBio.02371-21",
"doi-asserted-by": "crossref",
"key": "ref_15",
"unstructured": "Bracquemond, D., and Muriaux, D. (2021). Betacoronavirus Assembly: Clues and Perspectives for Elucidating SARS-CoV-2 Particle Formation and Egress. mBio, 12."
},
{
"DOI": "10.1016/S0065-3527(05)64006-7",
"article-title": "Molecular Interactions in the Assembly of Coronaviruses",
"author": "Rottier",
"doi-asserted-by": "crossref",
"first-page": "165",
"journal-title": "Advances in Virus Research",
"key": "ref_16",
"volume": "Volume 64",
"year": "2005"
},
{
"DOI": "10.1128/JVI.01052-08",
"article-title": "The M, E, and N Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus Are Required for Efficient Assembly, Trafficking, and Release of Virus-Like Particles",
"author": "Siu",
"doi-asserted-by": "crossref",
"first-page": "11318",
"journal-title": "J. Virol.",
"key": "ref_17",
"volume": "82",
"year": "2008"
},
{
"DOI": "10.1038/s41580-021-00418-x",
"article-title": "Mechanisms of SARS-CoV-2 Entry into Cells",
"author": "Jackson",
"doi-asserted-by": "crossref",
"first-page": "3",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_18",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"article-title": "A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "270",
"journal-title": "Nature",
"key": "ref_19",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"article-title": "SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "271",
"journal-title": "Cell",
"key": "ref_20",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.jbc.2021.100306",
"doi-asserted-by": "crossref",
"key": "ref_21",
"unstructured": "Bayati, A., Kumar, R., Francis, V., and McPherson, P.S. (2021). SARS-CoV-2 Infects Cells after Viral Entry via Clathrin-Mediated Endocytosis. J. Biol. Chem., 296."
},
{
"DOI": "10.1073/pnas.2003138117",
"article-title": "Cell Entry Mechanisms of SARS-CoV-2",
"author": "Shang",
"doi-asserted-by": "crossref",
"first-page": "11727",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_22",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.2147/HMER.S301979",
"article-title": "Binding of the SARS-CoV-2 Spike Protein to the Asialoglycoprotein Receptor on Human Primary Hepatocytes and Immortalized Hepatocyte-Like Cells by Confocal Analysis",
"author": "Collins",
"doi-asserted-by": "crossref",
"first-page": "37",
"journal-title": "Hepatic Med. Evid. Res.",
"key": "ref_23",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1038/s41422-021-00595-6",
"article-title": "Receptome Profiling Identifies KREMEN1 and ASGR1 as Alternative Functional Receptors of SARS-CoV-2",
"author": "Gu",
"doi-asserted-by": "crossref",
"first-page": "24",
"journal-title": "Cell Res.",
"key": "ref_24",
"volume": "32",
"year": "2022"
},
{
"DOI": "10.1016/j.immuni.2021.05.006",
"article-title": "SARS-CoV-2 Exacerbates Proinflammatory Responses in Myeloid Cells through C-Type Lectin Receptors and Tweety Family Member 2",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "1304",
"journal-title": "Immunity",
"key": "ref_25",
"volume": "54",
"year": "2021"
},
{
"DOI": "10.1038/s42255-020-00324-0",
"article-title": "HDL-Scavenger Receptor B Type 1 Facilitates SARS-CoV-2 Entry",
"author": "Wei",
"doi-asserted-by": "crossref",
"first-page": "1391",
"journal-title": "Nat. Metab.",
"key": "ref_26",
"volume": "2",
"year": "2020"
},
{
"DOI": "10.1021/acschemneuro.0c00619",
"article-title": "Novel Compounds Targeting Neuropilin Receptor 1 with Potential To Interfere with SARS-CoV-2 Virus Entry",
"author": "Patek",
"doi-asserted-by": "crossref",
"first-page": "1299",
"journal-title": "ACS Chem. Neurosci.",
"key": "ref_27",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1002/jmv.27244",
"article-title": "SARS-CoV-2 Pseudovirus Infectivity and Expression of Viral Entry-related Factors ACE2, TMPRSS2, Kim-1, and NRP-1 in Human Cells from the Respiratory, Urinary, Digestive, Reproductive, and Immune Systems",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "6671",
"journal-title": "J. Med. Virol.",
"key": "ref_28",
"volume": "93",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2023.06.005",
"article-title": "TMEM106B Is a Receptor Mediating ACE2-Independent SARS-CoV-2 Cell Entry",
"author": "Baggen",
"doi-asserted-by": "crossref",
"first-page": "3427",
"journal-title": "Cell",
"key": "ref_29",
"volume": "186",
"year": "2023"
},
{
"DOI": "10.1073/pnas.2317026121",
"article-title": "Human Transferrin Receptor Can Mediate SARS-CoV-2 Infection",
"author": "Liao",
"doi-asserted-by": "crossref",
"first-page": "e2317026121",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_30",
"volume": "121",
"year": "2024"
},
{
"DOI": "10.1038/s41467-023-44504-4",
"article-title": "Tubeimosides Are Pan-Coronavirus and Filovirus Inhibitors That Can Block Their Fusion Protein Binding to Niemann-Pick C1",
"author": "Khan",
"doi-asserted-by": "crossref",
"first-page": "162",
"journal-title": "Nat. Commun.",
"key": "ref_31",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1101/2020.03.24.005298",
"doi-asserted-by": "crossref",
"key": "ref_32",
"unstructured": "Snijder, E.J., Limpens, R.W.A.L., de Wilde, A.H., de Jong, A.W.M., Zevenhoven-Dobbe, J.C., Maier, H.J., Faas, F.F.G.A., Koster, A.J., and Bárcena, M. (2020). A Unifying Structural and Functional Model of the Coronavirus Replication Organelle: Tracking down RNA Synthesis. PLoS Biol., 18."
},
{
"DOI": "10.1016/j.chom.2020.11.003",
"article-title": "Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies",
"author": "Cortese",
"doi-asserted-by": "crossref",
"first-page": "853",
"journal-title": "Cell Host Microbe",
"key": "ref_33",
"volume": "28",
"year": "2020"
},
{
"DOI": "10.1007/s00018-022-04469-x",
"article-title": "The Double-Membrane Vesicle (DMV): A Virus-Induced Organelle Dedicated to the Replication of SARS-CoV-2 and Other Positive-Sense Single-Stranded RNA Viruses",
"author": "Roingeard",
"doi-asserted-by": "crossref",
"first-page": "425",
"journal-title": "Cell. Mol. Life Sci. CMLS",
"key": "ref_34",
"volume": "79",
"year": "2022"
},
{
"DOI": "10.12703/r/10-17",
"article-title": "Membrane Remodeling by SARS-CoV-2 - Double-Enveloped Viral Replication",
"author": "Mohan",
"doi-asserted-by": "crossref",
"first-page": "17",
"journal-title": "Fac. Rev.",
"key": "ref_35",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.1038/s41467-020-19619-7",
"article-title": "SARS-CoV-2 Structure and Replication Characterized by in Situ Cryo-Electron Tomography",
"author": "Klein",
"doi-asserted-by": "crossref",
"first-page": "5885",
"journal-title": "Nat. Commun.",
"key": "ref_36",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1126/science.abd3629",
"article-title": "A Molecular Pore Spans the Double Membrane of the Coronavirus Replication Organelle",
"author": "Wolff",
"doi-asserted-by": "crossref",
"first-page": "1395",
"journal-title": "Science",
"key": "ref_37",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1016/bs.aivir.2016.08.008",
"article-title": "The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing",
"author": "Snijder",
"doi-asserted-by": "crossref",
"first-page": "59",
"journal-title": "Adv. Virus Res.",
"key": "ref_38",
"volume": "96",
"year": "2016"
},
{
"DOI": "10.1038/s41586-022-04835-6",
"article-title": "The Role of NSP6 in the Biogenesis of the SARS-CoV-2 Replication Organelle",
"author": "Ricciardi",
"doi-asserted-by": "crossref",
"first-page": "761",
"journal-title": "Nature",
"key": "ref_39",
"volume": "606",
"year": "2022"
},
{
"DOI": "10.1038/s41467-023-43666-5",
"article-title": "SARS-CoV-2 Nsp3 and Nsp4 Are Minimal Constituents of a Pore Spanning Replication Organelle",
"author": "Zimmermann",
"doi-asserted-by": "crossref",
"first-page": "7894",
"journal-title": "Nat. Commun.",
"key": "ref_40",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/j.cell.2020.05.034",
"article-title": "Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "417",
"journal-title": "Cell",
"key": "ref_41",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-19770-1",
"article-title": "Architecture of a SARS-CoV-2 Mini Replication and Transcription Complex",
"author": "Yan",
"doi-asserted-by": "crossref",
"first-page": "5874",
"journal-title": "Nat. Commun.",
"key": "ref_42",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2021.05.033",
"article-title": "Coupling of N7-Methyltransferase and 3′-5′ Exoribonuclease with SARS-CoV-2 Polymerase Reveals Mechanisms for Capping and Proofreading",
"author": "Yan",
"doi-asserted-by": "crossref",
"first-page": "3474",
"journal-title": "Cell",
"key": "ref_43",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41586-024-07817-y",
"article-title": "Molecular Architecture of Coronavirus Double-Membrane Vesicle Pore Complex",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "224",
"journal-title": "Nature",
"key": "ref_44",
"volume": "633",
"year": "2024"
},
{
"DOI": "10.1016/j.cell.2020.10.039",
"article-title": "β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway",
"author": "Ghosh",
"doi-asserted-by": "crossref",
"first-page": "1520",
"journal-title": "Cell",
"key": "ref_45",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1038/s41467-021-24887-y",
"article-title": "Correlative Multi-Scale Cryo-Imaging Unveils SARS-CoV-2 Assembly and Egress",
"author": "Howe",
"doi-asserted-by": "crossref",
"first-page": "4629",
"journal-title": "Nat. Commun.",
"key": "ref_46",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1126/sciadv.abl4895",
"article-title": "SARS-CoV-2 Nucleocapsid Protein Adheres to Replication Organelles before Viral Assembly at the Golgi/ERGIC and Lysosome-Mediated Egress",
"author": "Scherer",
"doi-asserted-by": "crossref",
"first-page": "eabl4895",
"journal-title": "Sci. Adv.",
"key": "ref_47",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.3389/fbioe.2020.00862",
"doi-asserted-by": "crossref",
"key": "ref_48",
"unstructured": "Xu, R., Shi, M., Li, J., Song, P., and Li, N. (2020). Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol., 8."
},
{
"DOI": "10.1074/jbc.RA120.016148",
"doi-asserted-by": "crossref",
"key": "ref_49",
"unstructured": "Plescia, C.B., David, E.A., Patra, D., Sengupta, R., Amiar, S., Su, Y., and Stahelin, R.V. (2021). SARS-CoV-2 Viral Budding and Entry Can Be Modeled Using BSL-2 Level Virus-like Particles. J. Biol. Chem., 296."
},
{
"DOI": "10.3390/v14122825",
"doi-asserted-by": "crossref",
"key": "ref_50",
"unstructured": "Chang, Y.-S., Chu, L.-W., Chen, Z.-Y., Wu, J.-S., Su, W.-C., Yang, C.-J., Ping, Y.-H., and Lin, C.-W. (2022). Development of Fluorescence-Tagged SARS-CoV-2 Virus-like Particles by a Tri-Cistronic Vector Expression System for Investigating the Cellular Entry of SARS-CoV-2. Viruses, 14."
},
{
"DOI": "10.1126/science.abl6184",
"article-title": "Rapid Assessment of SARS-CoV-2–Evolved Variants Using Virus-like Particles",
"author": "Syed",
"doi-asserted-by": "crossref",
"first-page": "1626",
"journal-title": "Science",
"key": "ref_51",
"volume": "374",
"year": "2021"
},
{
"DOI": "10.1038/s41467-020-20768-y",
"article-title": "The SARS-CoV-2 Nucleocapsid Phosphoprotein Forms Mutually Exclusive Condensates with RNA and the Membrane-Associated M Protein",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "502",
"journal-title": "Nat. Commun.",
"key": "ref_52",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1021/acsomega.3c01625",
"article-title": "In Vitro Reconstitution and Analysis of SARS-CoV-2/Host Protein–Protein Interactions",
"author": "Moradi",
"doi-asserted-by": "crossref",
"first-page": "25009",
"journal-title": "ACS Omega",
"key": "ref_53",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.1101/2022.05.23.493138",
"doi-asserted-by": "crossref",
"key": "ref_54",
"unstructured": "Carlson, C.R., Adly, A.N., Bi, M., Howard, C.J., Frost, A., Cheng, Y., and Morgan, D.O. (2022). Reconstitution of the SARS-CoV-2 Ribonucleosome Provides Insights into Genomic RNA Packaging and Regulation by Phosphorylation. J. Biol. Chem., 298."
},
{
"DOI": "10.1021/acsnano.3c07265",
"article-title": "Three-Dimensional Remodeling of SARS-CoV2-Infected Cells Revealed by Cryogenic Soft X-Ray Tomography",
"author": "Castro",
"doi-asserted-by": "crossref",
"first-page": "22708",
"journal-title": "ACS Nano",
"key": "ref_55",
"volume": "17",
"year": "2023"
},
{
"DOI": "10.3390/v14122786",
"doi-asserted-by": "crossref",
"key": "ref_56",
"unstructured": "Bergner, T., Zech, F., Hirschenberger, M., Stenger, S., Sparrer, K.M.J., Kirchhoff, F., and Read, C. (2022). Near-Native Visualization of SARS-CoV-2 Induced Membrane Remodeling and Virion Morphogenesis. Viruses, 14."
},
{
"DOI": "10.1039/D1FD00031D",
"article-title": "Molecular Interactions of the M and E Integral Membrane Proteins of SARS-CoV-2",
"author": "Voth",
"doi-asserted-by": "crossref",
"first-page": "49",
"journal-title": "Faraday Discuss.",
"key": "ref_57",
"volume": "232",
"year": "2021"
},
{
"DOI": "10.1016/j.bbrep.2021.101023",
"doi-asserted-by": "crossref",
"key": "ref_58",
"unstructured": "Aldaais, E.A., Yegnaswamy, S., Albahrani, F., Alsowaiket, F., and Alramadan, S. (2021). Sequence and Structural Analysis of COVID-19 E and M Proteins with MERS Virus E and M Proteins—A Comparative Study. Biochem. Biophys. Rep., 26."
},
{
"DOI": "10.1038/s41467-024-44958-0",
"article-title": "Proteomic Analysis of SARS-CoV-2 Particles Unveils a Key Role of G3BP Proteins in Viral Assembly",
"author": "Murigneux",
"doi-asserted-by": "crossref",
"first-page": "640",
"journal-title": "Nat. Commun.",
"key": "ref_59",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1080/07391102.2020.1861983",
"article-title": "Structure and Dynamics of Membrane Protein in SARS-CoV-2",
"author": "Mahtarin",
"doi-asserted-by": "crossref",
"first-page": "4725",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "ref_60",
"volume": "40",
"year": "2022"
},
{
"DOI": "10.1155/2020/4389089",
"doi-asserted-by": "crossref",
"key": "ref_61",
"unstructured": "Bianchi, M., Benvenuto, D., Giovanetti, M., Angeletti, S., Ciccozzi, M., and Pascarella, S. (2020). Sars-CoV-2 Envelope and Membrane Proteins: Structural Differences Linked to Virus Characteristics?. BioMed Res. Int., 2020."
},
{
"DOI": "10.1038/s41467-022-32019-3",
"article-title": "Structure of SARS-CoV-2 Membrane Protein Essential for Virus Assembly",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "4399",
"journal-title": "Nat. Commun.",
"key": "ref_62",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1128/JVI.01131-10",
"article-title": "A Conserved Domain in the Coronavirus Membrane Protein Tail Is Important for Virus Assembly",
"author": "Arndt",
"doi-asserted-by": "crossref",
"first-page": "11418",
"journal-title": "J. Virol.",
"key": "ref_63",
"volume": "84",
"year": "2010"
},
{
"DOI": "10.1007/s00018-023-05008-y",
"article-title": "The KxGxYR and DxE Motifs in the C-Tail of the Middle East Respiratory Syndrome Coronavirus Membrane Protein Are Crucial for Infectious Virus Assembly",
"author": "Desmarets",
"doi-asserted-by": "crossref",
"first-page": "353",
"journal-title": "Cell. Mol. Life Sci. CMLS",
"key": "ref_64",
"volume": "80",
"year": "2023"
},
{
"DOI": "10.1074/jbc.RA120.016175",
"doi-asserted-by": "crossref",
"key": "ref_65",
"unstructured": "Boson, B., Legros, V., Zhou, B., Siret, E., Mathieu, C., Cosset, F.-L., Lavillette, D., and Denolly, S. (2021). The SARS-CoV-2 Envelope and Membrane Proteins Modulate Maturation and Retention of the Spike Protein, Allowing Assembly of Virus-like Particles. J. Biol. Chem., 296."
},
{
"DOI": "10.1080/07391102.2021.2016490",
"article-title": "An Insight into SARS-CoV-2 Membrane Protein Interaction with Spike, Envelope, and Nucleocapsid Proteins",
"author": "Kumar",
"doi-asserted-by": "crossref",
"first-page": "1062",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "ref_66",
"volume": "41",
"year": "2023"
},
{
"DOI": "10.1128/JVI.73.9.7441-7452.1999",
"article-title": "Mapping of the Coronavirus Membrane Protein Domains Involved in Interaction with the Spike Protein",
"author": "Smeets",
"doi-asserted-by": "crossref",
"first-page": "7441",
"journal-title": "J. Virol.",
"key": "ref_67",
"volume": "73",
"year": "1999"
},
{
"DOI": "10.1098/rsob.200209",
"doi-asserted-by": "crossref",
"key": "ref_68",
"unstructured": "Duart, G., García-Murria, M.J., Grau, B., Acosta-Cáceres, J.M., Martínez-Gil, L., and Mingarro, I. (2020). SARS-CoV-2 Envelope Protein Topology in Eukaryotic Membranes. Open Biol., 10."
},
{
"DOI": "10.1016/j.virol.2012.07.005",
"article-title": "Coronavirus E Protein Forms Ion Channels with Functionally and Structurally-Involved Membrane Lipids",
"author": "Alcaraz",
"doi-asserted-by": "crossref",
"first-page": "485",
"journal-title": "Virology",
"key": "ref_69",
"volume": "432",
"year": "2012"
},
{
"DOI": "10.1038/s42003-023-05490-x",
"doi-asserted-by": "crossref",
"key": "ref_70",
"unstructured": "Zhang, R., Qin, H., Prasad, R., Fu, R., Zhou, H.-X., and Cross, T.A. (2023). Dimeric Transmembrane Structure of the SARS-CoV-2 E Protein. Commun. Biol., 6."
},
{
"DOI": "10.1021/acs.langmuir.3c03079",
"article-title": "Membrane Condensation and Curvature Induced by SARS-CoV-2 Envelope Protein",
"author": "Shen",
"doi-asserted-by": "crossref",
"first-page": "2646",
"journal-title": "Langmuir",
"key": "ref_71",
"volume": "40",
"year": "2024"
},
{
"DOI": "10.1242/jcs.260685",
"article-title": "SARS-CoV-2 Infection Alkalinizes the ERGIC and Lysosomes through the Viroporin Activity of the Viral Envelope Protein",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "jcs260685",
"journal-title": "J. Cell Sci.",
"key": "ref_72",
"volume": "136",
"year": "2023"
},
{
"DOI": "10.1126/sciadv.adl5012",
"article-title": "ER-Export and ARFRP1/AP-1-Dependent Delivery of SARS-CoV-2 Envelope to Lysosomes Controls Late Stages of Viral Replication",
"author": "Pearson",
"doi-asserted-by": "crossref",
"first-page": "eadl5012",
"journal-title": "Sci. Adv.",
"key": "ref_73",
"volume": "10",
"year": "2024"
},
{
"DOI": "10.1038/s41594-020-00536-8",
"article-title": "Structure and Drug Binding of the SARS-CoV-2 Envelope Protein Transmembrane Domain in Lipid Bilayers",
"author": "Mandala",
"doi-asserted-by": "crossref",
"first-page": "1202",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "ref_74",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1101/2023.03.29.534854",
"doi-asserted-by": "crossref",
"key": "ref_75",
"unstructured": "Ewart, G., Bobardt, M., Bentzen, B.H., Yan, Y., Thomson, A., Klumpp, K., Becker, S., Rosenkilde, M.M., Miller, M., and Gallay, P. (2023). Post-Infection Treatment with the E Protein Inhibitor BIT225 Reduces Disease Severity and Increases Survival of K18-hACE2 Transgenic Mice Infected with a Lethal Dose of SARS-CoV-2. PLoS Pathog., 19."
},
{
"DOI": "10.1126/sciadv.adi9007",
"article-title": "Atomic Structure of the Open SARS-CoV-2 E Viroporin",
"author": "Dregni",
"doi-asserted-by": "crossref",
"first-page": "eadi9007",
"journal-title": "Sci. Adv.",
"key": "ref_76",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1002/prot.26317",
"article-title": "Structure and Dynamics of the SARS-CoV-2 Envelope Protein Monomer",
"author": "Kuzmin",
"doi-asserted-by": "crossref",
"first-page": "1102",
"journal-title": "Proteins",
"key": "ref_77",
"volume": "90",
"year": "2022"
},
{
"DOI": "10.1038/srep44695",
"doi-asserted-by": "crossref",
"key": "ref_78",
"unstructured": "Martyna, A., Bahsoun, B., Badham, M.D., Srinivasan, S., Howard, M.J., and Rossman, J.S. (2017). Membrane Remodeling by the M2 Amphipathic Helix Drives Influenza Virus Membrane Scission. Sci. Rep., 7."
},
{
"DOI": "10.1016/j.cell.2010.08.029",
"article-title": "Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission",
"author": "Rossman",
"doi-asserted-by": "crossref",
"first-page": "902",
"journal-title": "Cell",
"key": "ref_79",
"volume": "142",
"year": "2010"
},
{
"DOI": "10.1038/s41467-021-21953-3",
"article-title": "The SARS-CoV-2 Nucleocapsid Protein Is Dynamic, Disordered, and Phase Separates with RNA",
"author": "Cubuk",
"doi-asserted-by": "crossref",
"first-page": "1936",
"journal-title": "Nat. Commun.",
"key": "ref_80",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.15252/embj.2020105938",
"article-title": "Structures of the SARS -CoV-2 Nucleocapsid and Their Perspectives for Drug Design",
"author": "Peng",
"doi-asserted-by": "crossref",
"first-page": "e105938",
"journal-title": "EMBO J.",
"key": "ref_81",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.09.018",
"article-title": "Molecular Architecture of the SARS-CoV-2 Virus",
"author": "Yao",
"doi-asserted-by": "crossref",
"first-page": "730",
"journal-title": "Cell",
"key": "ref_82",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-19843-1",
"article-title": "Nucleocapsid Protein of SARS-CoV-2 Phase Separates into RNA-Rich Polymerase-Containing Condensates",
"author": "Savastano",
"doi-asserted-by": "crossref",
"first-page": "6041",
"journal-title": "Nat. Commun.",
"key": "ref_83",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1371/journal.pbio.3001425",
"doi-asserted-by": "crossref",
"key": "ref_84",
"unstructured": "Jack, A., Ferro, L.S., Trnka, M.J., Wehri, E., Nadgir, A., Nguyenla, X., Fox, D., Costa, K., Stanley, S., and Schaletzky, J. (2021). SARS-CoV-2 Nucleocapsid Protein Forms Condensates with Viral Genomic RNA. PLoS Biol., 19."
},
{
"DOI": "10.1038/s42003-024-06130-8",
"doi-asserted-by": "crossref",
"key": "ref_85",
"unstructured": "Du, L., Deiter, F., Bouzidi, M.S., Billaud, J.-N., Simmons, G., Dabral, P., Selvarajah, S., Lingappa, A.F., Michon, M., and Yu, S.F. (2024). A Viral Assembly Inhibitor Blocks SARS-CoV-2 Replication in Airway Epithelial Cells. Commun. Biol., 7."
},
{
"DOI": "10.1016/j.chom.2020.08.004",
"article-title": "Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "586",
"journal-title": "Cell Host Microbe",
"key": "ref_86",
"volume": "28",
"year": "2020"
},
{
"DOI": "10.1016/S0065-3527(06)66005-3",
"article-title": "The Molecular Biology of Coronaviruses",
"author": "Masters",
"doi-asserted-by": "crossref",
"first-page": "193",
"journal-title": "Advances in Virus Research",
"key": "ref_87",
"volume": "Volume 66",
"year": "2006"
},
{
"DOI": "10.1126/science.abb2507",
"article-title": "Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation",
"author": "Wrapp",
"doi-asserted-by": "crossref",
"first-page": "1260",
"journal-title": "Science",
"key": "ref_88",
"volume": "367",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.02.058",
"article-title": "Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein",
"author": "Walls",
"doi-asserted-by": "crossref",
"first-page": "281",
"journal-title": "Cell",
"key": "ref_89",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-19808-4",
"article-title": "SARS-CoV-2 Spike-Protein D614G Mutation Increases Virion Spike Density and Infectivity",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "6013",
"journal-title": "Nat. Commun.",
"key": "ref_90",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.scr.2020.102115",
"article-title": "Furin Cleavage Sites Naturally Occur in Coronaviruses",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "102115",
"journal-title": "Stem Cell Res.",
"key": "ref_91",
"volume": "50",
"year": "2020"
},
{
"DOI": "10.3390/v13010109",
"doi-asserted-by": "crossref",
"key": "ref_92",
"unstructured": "Xia, X. (2021). Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses, 13."
},
{
"DOI": "10.1038/s41467-021-25589-1",
"article-title": "Sequences in the Cytoplasmic Tail of SARS-CoV-2 Spike Facilitate Expression at the Cell Surface and Syncytia Formation",
"author": "Welch",
"doi-asserted-by": "crossref",
"first-page": "5333",
"journal-title": "Nat. Commun.",
"key": "ref_93",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41392-021-00809-8",
"article-title": "The Glycosylation in SARS-CoV-2 and Its Receptor ACE2",
"author": "Gong",
"doi-asserted-by": "crossref",
"first-page": "396",
"journal-title": "Signal Transduct. Target. Ther.",
"key": "ref_94",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1128/mbio.01672-23",
"article-title": "SARS-CoV-2 Spike Glycosylation Affects Function and Neutralization Sensitivity",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "e01672-23",
"journal-title": "mBio",
"key": "ref_95",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1371/journal.ppat.1011788",
"doi-asserted-by": "crossref",
"key": "ref_96",
"unstructured": "Lusvarghi, S., Stauft, C.B., Vassell, R., Williams, B., Baha, H., Wang, W., Neerukonda, S.N., Wang, T., and Weiss, C.D. (2023). Effects of N-Glycan Modifications on Spike Expression, Virus Infectivity, and Neutralization Sensitivity in Ancestral Compared to Omicron SARS-CoV-2 Variants. PLoS Pathog., 19."
},
{
"DOI": "10.1128/JVI.79.22.13848-13855.2005",
"article-title": "Assembly of Severe Acute Respiratory Syndrome Coronavirus RNA Packaging Signal into Virus-Like Particles Is Nucleocapsid Dependent",
"author": "Hsieh",
"doi-asserted-by": "crossref",
"first-page": "13848",
"journal-title": "J. Virol.",
"key": "ref_97",
"volume": "79",
"year": "2005"
},
{
"DOI": "10.1002/1873-3468.14109",
"article-title": "A Weak COPI Binding Motif in the Cytoplasmic Tail of SARS-CoV-2 Spike Glycoprotein Is Necessary for Its Cleavage, Glycosylation, and Localization",
"author": "Jennings",
"doi-asserted-by": "crossref",
"first-page": "1758",
"journal-title": "FEBS Lett.",
"key": "ref_98",
"volume": "595",
"year": "2021"
},
{
"DOI": "10.1128/JVI.02146-06",
"article-title": "The Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Contains a Novel Endoplasmic Reticulum Retrieval Signal That Binds COPI and Promotes Interaction with Membrane Protein",
"author": "McBride",
"doi-asserted-by": "crossref",
"first-page": "2418",
"journal-title": "J. Virol.",
"key": "ref_99",
"volume": "81",
"year": "2007"
},
{
"DOI": "10.1186/s12977-015-0197-x",
"article-title": "Evidence That the Endosomal Sorting Complex Required for Transport-II (ESCRT-II) Is Required for Efficient Human Immunodeficiency Virus-1 (HIV-1) Production",
"author": "Meng",
"doi-asserted-by": "crossref",
"first-page": "72",
"journal-title": "Retrovirology",
"key": "ref_100",
"volume": "12",
"year": "2015"
},
{
"DOI": "10.1073/pnas.131059198",
"article-title": "Tsg101, a Homologue of Ubiquitin-Conjugating (E2) Enzymes, Binds the L Domain in HIV Type 1 Pr55 Gag",
"author": "VerPlank",
"doi-asserted-by": "crossref",
"first-page": "7724",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_101",
"volume": "98",
"year": "2001"
},
{
"DOI": "10.1016/S0092-8674(01)00506-2",
"article-title": "Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding",
"author": "Garrus",
"doi-asserted-by": "crossref",
"first-page": "55",
"journal-title": "Cell",
"key": "ref_102",
"volume": "107",
"year": "2001"
},
{
"DOI": "10.1038/nm1201-1313",
"article-title": "HIV-1 and Ebola Virus Encode Small Peptide Motifs That Recruit Tsg101 to Sites of Particle Assembly to Facilitate Egress",
"author": "Zang",
"doi-asserted-by": "crossref",
"first-page": "1313",
"journal-title": "Nat. Med.",
"key": "ref_103",
"volume": "7",
"year": "2001"
},
{
"DOI": "10.1073/pnas.032511899",
"article-title": "Overexpression of the N-Terminal Domain of TSG101 Inhibits HIV-1 Budding by Blocking Late Domain Function",
"author": "Demirov",
"doi-asserted-by": "crossref",
"first-page": "955",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_104",
"volume": "99",
"year": "2002"
},
{
"DOI": "10.1128/JVI.76.10.4679-4687.2002",
"article-title": "Viral Late Domains",
"author": "Freed",
"doi-asserted-by": "crossref",
"first-page": "4679",
"journal-title": "J. Virol.",
"key": "ref_105",
"volume": "76",
"year": "2002"
},
{
"DOI": "10.3390/v13081559",
"doi-asserted-by": "crossref",
"key": "ref_106",
"unstructured": "Welker, L., Paillart, J.-C., and Bernacchi, S. (2021). Importance of Viral Late Domains in Budding and Release of Enveloped RNA Viruses. Viruses, 13."
},
{
"DOI": "10.1038/s41580-019-0177-4",
"article-title": "The Many Functions of ESCRTs",
"author": "Vietri",
"doi-asserted-by": "crossref",
"first-page": "25",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_107",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1038/nature07961",
"article-title": "The ESCRT Machinery in Endosomal Sorting of Ubiquitylated Membrane Proteins",
"author": "Raiborg",
"doi-asserted-by": "crossref",
"first-page": "445",
"journal-title": "Nature",
"key": "ref_108",
"volume": "458",
"year": "2009"
},
{
"DOI": "10.15252/embj.201592484",
"article-title": "ESCRT s Are Everywhere",
"author": "Hurley",
"doi-asserted-by": "crossref",
"first-page": "2398",
"journal-title": "EMBO J.",
"key": "ref_109",
"volume": "34",
"year": "2015"
},
{
"DOI": "10.1038/nrm.2016.121",
"article-title": "Reverse-Topology Membrane Scission by the ESCRT Proteins",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "5",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_110",
"volume": "18",
"year": "2017"
},
{
"DOI": "10.1128/jvi.00426-23",
"article-title": "Distinct Motifs in the E Protein Are Required for SARS-CoV-2 Virus Particle Formation and Lysosomal Deacidification in Host Cells",
"author": "Miura",
"doi-asserted-by": "crossref",
"first-page": "e00426-23",
"journal-title": "J. Virol.",
"key": "ref_111",
"volume": "97",
"year": "2023"
},
{
"DOI": "10.1128/JVI.01605-19",
"article-title": "Amphipathic Helices of Cellular Proteins Can Replace the Helix in M2 of Influenza A Virus with Only Small Effects on Virus Replication",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "e01605-19",
"journal-title": "J. Virol.",
"key": "ref_112",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1021/ja400146z",
"article-title": "Influenza Virus A M2 Protein Generates Negative Gaussian Membrane Curvature Necessary for Budding and Scission",
"author": "Schmidt",
"doi-asserted-by": "crossref",
"first-page": "13710",
"journal-title": "J. Am. Chem. Soc.",
"key": "ref_113",
"volume": "135",
"year": "2013"
},
{
"DOI": "10.3389/fcell.2019.00291",
"doi-asserted-by": "crossref",
"key": "ref_114",
"unstructured": "Zhukovsky, M.A., Filograna, A., Luini, A., Corda, D., and Valente, C. (2019). Protein Amphipathic Helix Insertion: A Mechanism to Induce Membrane Fission. Front. Cell Dev. Biol., 7."
},
{
"DOI": "10.1038/sj.emboj.7601174",
"article-title": "Mechanism of Endophilin N-BAR Domain-Mediated Membrane Curvature",
"author": "Gallop",
"doi-asserted-by": "crossref",
"first-page": "2898",
"journal-title": "EMBO J.",
"key": "ref_115",
"volume": "25",
"year": "2006"
},
{
"DOI": "10.1016/j.biocel.2006.12.004",
"article-title": "Epsin: Inducing Membrane Curvature",
"author": "Horvath",
"doi-asserted-by": "crossref",
"first-page": "1765",
"journal-title": "Int. J. Biochem. Cell Biol.",
"key": "ref_116",
"volume": "39",
"year": "2007"
},
{
"DOI": "10.3390/v12091054",
"doi-asserted-by": "crossref",
"key": "ref_117",
"unstructured": "Alsaadi, E.A.J., Neuman, B.W., and Jones, I.M. (2020). Identification of a Membrane Binding Peptide in the Envelope Protein of MHV Coronavirus. Viruses, 12."
},
{
"DOI": "10.1006/viro.1999.9955",
"article-title": "Release of Coronavirus E Protein in Membrane Vesicles from Virus-Infected Cells and E Protein-Expressing Cells",
"author": "Maeda",
"doi-asserted-by": "crossref",
"first-page": "265",
"journal-title": "Virology",
"key": "ref_118",
"volume": "263",
"year": "1999"
},
{
"DOI": "10.1038/s41421-023-00575-7",
"article-title": "Impact of SARS-CoV-2 Envelope Protein Mutations on the Pathogenicity of Omicron XBB",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "80",
"journal-title": "Cell Discov.",
"key": "ref_119",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1186/s12985-022-01951-7",
"article-title": "Mutations in SARS-CoV-2 Structural Proteins: A Global Analysis",
"author": "Abavisani",
"doi-asserted-by": "crossref",
"first-page": "220",
"journal-title": "Virol. J.",
"key": "ref_120",
"volume": "19",
"year": "2022"
},
{
"DOI": "10.3390/cells10040853",
"doi-asserted-by": "crossref",
"key": "ref_121",
"unstructured": "Kumar, B., Hawkins, G.M., Kicmal, T., Qing, E., Timm, E., and Gallagher, T. (2021). Assembly and Entry of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV2): Evaluation Using Virus-Like Particles. Cells, 10."
},
{
"DOI": "10.1038/s41598-022-18681-z",
"doi-asserted-by": "crossref",
"key": "ref_122",
"unstructured": "Gourdelier, M., Swain, J., Arone, C., Mouttou, A., Bracquemond, D., Merida, P., Saffarian, S., Lyonnais, S., Favard, C., and Muriaux, D. (2022). Optimized Production and Fluorescent Labeling of SARS-CoV-2 Virus-like Particles. Sci. Rep., 12."
},
{
"DOI": "10.1016/j.devcel.2021.10.006",
"article-title": "ORF3a of SARS-CoV-2 Promotes Lysosomal Exocytosis-Mediated Viral Egress",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "3250",
"journal-title": "Dev. Cell",
"key": "ref_123",
"volume": "56",
"year": "2021"
},
{
"DOI": "10.1038/s41581-019-0129-4",
"article-title": "Glycosylation in Health and Disease",
"author": "Reily",
"doi-asserted-by": "crossref",
"first-page": "346",
"journal-title": "Nat. Rev. Nephrol.",
"key": "ref_124",
"volume": "15",
"year": "2019"
},
{
"DOI": "10.1021/acscentsci.1c00058",
"article-title": "Assessing Antigen Structural Integrity through Glycosylation Analysis of the SARS-CoV-2 Viral Spike",
"author": "Brun",
"doi-asserted-by": "crossref",
"first-page": "586",
"journal-title": "ACS Cent. Sci.",
"key": "ref_125",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.1016/S0161-5890(03)00155-X",
"article-title": "The Role of Mannose-Binding Lectin in Health and Disease",
"author": "Turner",
"doi-asserted-by": "crossref",
"first-page": "423",
"journal-title": "Mol. Immunol.",
"key": "ref_126",
"volume": "40",
"year": "2003"
},
{
"DOI": "10.1242/jcs.03483",
"article-title": "Poliovirus Infection Blocks ERGIC-to-Golgi Trafficking and Induces Microtubule-Dependent Disruption of the Golgi Complex",
"author": "Beske",
"doi-asserted-by": "crossref",
"first-page": "3207",
"journal-title": "J. Cell Sci.",
"key": "ref_127",
"volume": "120",
"year": "2007"
},
{
"DOI": "10.1083/jcb.99.3.1101",
"article-title": "Effect of Microtubule Assembly Status on the Intracellular Processing and Surface Expression of an Integral Protein of the Plasma Membrane",
"author": "Rogalski",
"doi-asserted-by": "crossref",
"first-page": "1101",
"journal-title": "J. Cell Biol.",
"key": "ref_128",
"volume": "99",
"year": "1984"
},
{
"DOI": "10.1091/mbc.7.4.631",
"article-title": "Golgi Dispersal during Microtubule Disruption: Regeneration of Golgi Stacks at Peripheral Endoplasmic Reticulum Exit Sites",
"author": "Cole",
"doi-asserted-by": "crossref",
"first-page": "631",
"journal-title": "Mol. Biol. Cell",
"key": "ref_129",
"volume": "7",
"year": "1996"
},
{
"DOI": "10.1101/cshperspect.a005181",
"doi-asserted-by": "crossref",
"key": "ref_130",
"unstructured": "Klumperman, J. (2011). Architecture of the Mammalian Golgi. Cold Spring Harb. Perspect. Biol., 3."
},
{
"DOI": "10.1101/2022.03.04.483074",
"doi-asserted-by": "crossref",
"key": "ref_131",
"unstructured": "Zhang, J., Kennedy, A., de Melo Jorge, D.M., Xing, L., Reid, W., Bui, S., Joppich, J., Rose, M., Ercan, S., and Tang, Q. (2024). SARS-CoV-2 Remodels the Golgi Apparatus to Facilitate Viral Assembly and Secretion. BioRxiv Prepr. Serv. Biol., 2022.03.04.483074."
},
{
"DOI": "10.1006/viro.1998.9032",
"article-title": "Virus-Induced Host Gene Shutoff in Animals and Plants",
"author": "Aranda",
"doi-asserted-by": "crossref",
"first-page": "261",
"journal-title": "Virology",
"key": "ref_132",
"volume": "243",
"year": "1998"
},
{
"DOI": "10.1038/s41586-021-03610-3",
"article-title": "SARS-CoV-2 Uses a Multipronged Strategy to Impede Host Protein Synthesis",
"author": "Finkel",
"doi-asserted-by": "crossref",
"first-page": "240",
"journal-title": "Nature",
"key": "ref_133",
"volume": "594",
"year": "2021"
},
{
"DOI": "10.1038/s41594-020-0511-8",
"article-title": "SARS-CoV-2 Nsp1 Binds the Ribosomal mRNA Channel to Inhibit Translation",
"author": "Schubert",
"doi-asserted-by": "crossref",
"first-page": "959",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "ref_134",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1126/science.abc8665",
"article-title": "Structural Basis for Translational Shutdown and Immune Evasion by the Nsp1 Protein of SARS-CoV-2",
"author": "Thoms",
"doi-asserted-by": "crossref",
"first-page": "1249",
"journal-title": "Science",
"key": "ref_135",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1073/pnas.2117198119",
"article-title": "Targeting Stem-Loop 1 of the SARS-CoV-2 5′ UTR to Suppress Viral Translation and Nsp1 Evasion",
"author": "Vora",
"doi-asserted-by": "crossref",
"first-page": "e2117198119",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_136",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.1016/j.isci.2022.104709",
"article-title": "Glycosylation and S-Palmitoylation Regulate SARS-CoV-2 Spike Protein Intracellular Trafficking",
"author": "Tien",
"doi-asserted-by": "crossref",
"first-page": "104709",
"journal-title": "iScience",
"key": "ref_137",
"volume": "25",
"year": "2022"
},
{
"DOI": "10.1038/s41392-021-00651-y",
"article-title": "Palmitoylation of SARS-CoV-2 S Protein Is Essential for Viral Infectivity",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "231",
"journal-title": "Signal Transduct. Target. Ther.",
"key": "ref_138",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1021/acs.jctc.1c00359",
"article-title": "Computational Study on the Function of Palmitoylation on the Envelope Protein in SARS-CoV-2",
"author": "Sun",
"doi-asserted-by": "crossref",
"first-page": "6483",
"journal-title": "J. Chem. Theory Comput.",
"key": "ref_139",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1016/j.jbc.2021.101112",
"doi-asserted-by": "crossref",
"key": "ref_140",
"unstructured": "Puthenveetil, R., Lun, C.M., Murphy, R.E., Healy, L.B., Vilmen, G., Christenson, E.T., Freed, E.O., and Banerjee, A. (2021). S-Acylation of SARS-CoV-2 Spike Protein: Mechanistic Dissection, in Vitro Reconstitution and Role in Viral Infectivity. J. Biol. Chem., 297."
},
{
"DOI": "10.1097/IM9.0000000000000071",
"article-title": "Mass Spectrometry Analysis of SARS-CoV-2 Nucleocapsid Protein Reveals Camouflaging Glycans and Unique Post-Translational Modifications",
"author": "Sun",
"doi-asserted-by": "crossref",
"first-page": "149",
"journal-title": "Infect. Microbes Dis.",
"key": "ref_141",
"volume": "3",
"year": "2021"
},
{
"DOI": "10.1016/j.jbc.2024.107354",
"doi-asserted-by": "crossref",
"key": "ref_142",
"unstructured": "Stuwe, H., Reardon, P.N., Yu, Z., Shah, S., Hughes, K., and Barbar, E.J. (2024). Phosphorylation in the Ser/Arg-Rich Region of the Nucleocapsid of SARS-CoV-2 Regulates Phase Separation by Inhibiting Self-Association of a Distant Helix. J. Biol. Chem."
},
{
"DOI": "10.1126/sciadv.aax2323",
"article-title": "A Specific Phosphorylation-Dependent Conformational Switch in SARS-CoV-2 Nucleocapsid Protein Inhibits RNA Binding",
"author": "Botova",
"doi-asserted-by": "crossref",
"first-page": "eaax2323",
"journal-title": "Sci. Adv.",
"key": "ref_143",
"volume": "10",
"year": "2024"
},
{
"DOI": "10.1016/j.jbc.2021.100821",
"doi-asserted-by": "crossref",
"key": "ref_144",
"unstructured": "Cai, T., Yu, Z., Wang, Z., Liang, C., and Richard, S. (2021). Arginine Methylation of SARS-CoV-2 Nucleocapsid Protein Regulates RNA Binding, Its Ability to Suppress Stress Granule Formation, and Viral Replication. J. Biol. Chem., 297."
},
{
"DOI": "10.1128/mbio.03168-21",
"article-title": "The E3 Ubiquitin Ligase RNF5 Facilitates SARS-CoV-2 Membrane Protein-Mediated Virion Release",
"author": "Yuan",
"doi-asserted-by": "crossref",
"first-page": "e03168-21",
"journal-title": "mBio",
"key": "ref_145",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.15252/msb.202110396",
"doi-asserted-by": "crossref",
"key": "ref_146",
"unstructured": "Liu, X., Huuskonen, S., Laitinen, T., Redchuk, T., Bogacheva, M., Salokas, K., Pöhner, I., Öhman, T., Tonduru, A.K., and Hassinen, A. (2021). SARS-CoV-2-Host Proteome Interactions for Antiviral Drug Discovery. Mol. Syst. Biol., 17."
},
{
"DOI": "10.1038/s41586-021-03493-4",
"article-title": "Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV",
"author": "Stukalov",
"doi-asserted-by": "crossref",
"first-page": "246",
"journal-title": "Nature",
"key": "ref_147",
"volume": "594",
"year": "2021"
},
{
"DOI": "10.1016/j.ijbiomac.2021.09.018",
"article-title": "Genomics-Guided Targeting of Stress Granule Proteins G3BP1/2 to Inhibit SARS-CoV-2 Propagation",
"author": "Ali",
"doi-asserted-by": "crossref",
"first-page": "636",
"journal-title": "Int. J. Biol. Macromol.",
"key": "ref_148",
"volume": "190",
"year": "2021"
},
{
"DOI": "10.1016/j.molcel.2021.04.008",
"article-title": "Functional Landscape of SARS-CoV-2 Cellular Restriction",
"author": "Lewinski",
"doi-asserted-by": "crossref",
"first-page": "2656",
"journal-title": "Mol. Cell",
"key": "ref_149",
"volume": "81",
"year": "2021"
},
{
"DOI": "10.1038/s41467-022-30609-9",
"article-title": "SARS-CoV-2 ORF7a Potently Inhibits the Antiviral Effect of the Host Factor SERINC5",
"author": "Timilsina",
"doi-asserted-by": "crossref",
"first-page": "2935",
"journal-title": "Nat. Commun.",
"key": "ref_150",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1128/mBio.01316-21",
"doi-asserted-by": "crossref",
"key": "ref_151",
"unstructured": "Wang, L., Sola, I., Enjuanes, L., and Zuñiga, S. (2021). MOV10 Helicase Interacts with Coronavirus Nucleocapsid Protein and Has Antiviral Activity. mBio, 12."
},
{
"DOI": "10.1073/pnas.2308355120",
"article-title": "Cellular Nucleic Acid-Binding Protein Restricts SARS-CoV-2 by Regulating Interferon and Disrupting RNA-Protein Condensates",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "e2308355120",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_152",
"volume": "120",
"year": "2023"
},
{
"DOI": "10.1038/s41587-022-01474-0",
"article-title": "A Comprehensive SARS-CoV-2-Human Protein-Protein Interactome Reveals COVID-19 Pathobiology and Potential Host Therapeutic Targets",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "128",
"journal-title": "Nat. Biotechnol.",
"key": "ref_153",
"volume": "41",
"year": "2023"
},
{
"DOI": "10.1101/2021.11.23.469747",
"doi-asserted-by": "crossref",
"key": "ref_154",
"unstructured": "Zhang, J., Li, Q., Cruz Cosme, R.S., Gerzanich, V., Tang, Q., Simard, J.M., and Zhao, R.Y. (2021). Genome-Wide Characterization of SARS-CoV-2 Cytopathogenic Proteins in the Search of Antiviral Targets. mBio, 13."
},
{
"DOI": "10.1128/JVI.02274-15",
"article-title": "Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference",
"author": "Taylor",
"doi-asserted-by": "crossref",
"first-page": "11820",
"journal-title": "J. Virol.",
"key": "ref_155",
"volume": "89",
"year": "2015"
}
],
"reference-count": 155,
"references-count": 155,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1999-4915/16/11/1648"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 Assembly: Gaining Infectivity and Beyond",
"type": "journal-article",
"volume": "16"
}